Corynebacterium glutamicum and method for overproduction of phosphatidylserine by means of corynebacterium glutamicum
A technology of phosphatidylserine and Corynebacterium glutamicum, which is applied in the fields of metabolic engineering and fermentation engineering, can solve the problems of low PS content, slow growth of yeast, difficulty in extracting high-purity PS, etc., and achieve the effect of increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
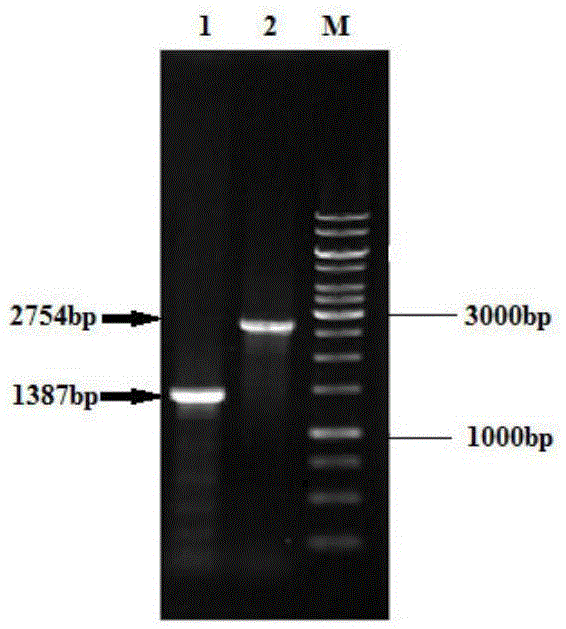
[0046] The construction of embodiment 1 Corynebacterium glutamicum sdaA deletion strain
[0047] (1) Construction of sdaA deletion plasmid
[0048] 1) Design two pairs of primers according to the sequence of the sdaA gene of Corynebacterium glutamicum in NCBI, and obtain the upstream 703bp fragment and the downstream 701bp fragment of the sdaA gene by PCR method, and design two pairs of primers as follows:
[0049] sdaAL-F: 5'— GGAATTCC ATTGCGGCATCTGCTGG—3'
[0050] sdaAL-R: 5'—CCGGTACCCGGGCTGAGGGTGGGCC—3'
[0051] sdaAR-F: 5'—CCCTCAGCCCGGTACGGCTTTAACACG—3'
[0052] sdaAR-R: 5'— GCTCTAGA GCTACACCGCTTGATCGCA—3'
[0053] 2) Using high-fidelity Pyrobest DNA polymerase, using the left and right homology arms as templates, the modified fragment △sdaA with the deletion of the sdaA gene was amplified by overlapping PCR;
[0054] 3) Ligate the recovered 1387bp target fragment ΔsdaA with pK18mobsacB treated with the same enzyme digestion to obtain pK18mobsacB ΔsdaA, and transfo...
Embodiment 2
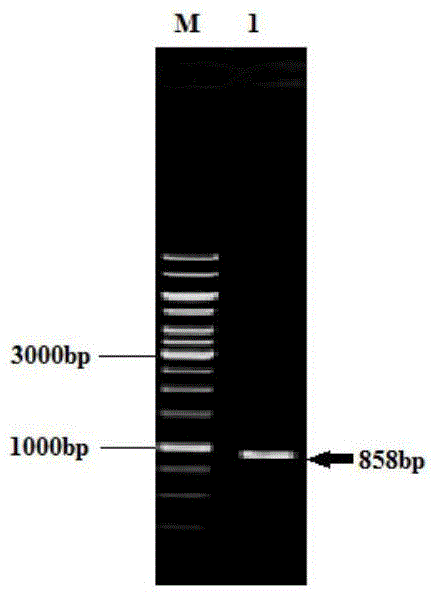
[0060] Example 2C. glutamicum(ΔsdaA) / pXMJ19-pssA-serA Δ197 - Construction of cdsA
[0061] 1. Expression plasmid pXMJ19-pssA-serA Δ197 - Construction of cdsA
[0062] 1) Using the PCR method to amplify the pssA gene using E.coliK12 genomic DNA as a template.
[0063] 2) The purified PCR product was double-digested with XbaI and EcoRI and connected to the expression vector pXMJ19 containing the same restriction site to construct the recombinant plasmid pXMJ19-pssA.
[0064] 3) The recombinant plasmid was transformed into E.coliJM109 competent cells, a single colony was picked and cultured on chloramphenicol-resistant LB medium, and the extracted plasmid was identified by double digestion with XbaI and EcoRI.
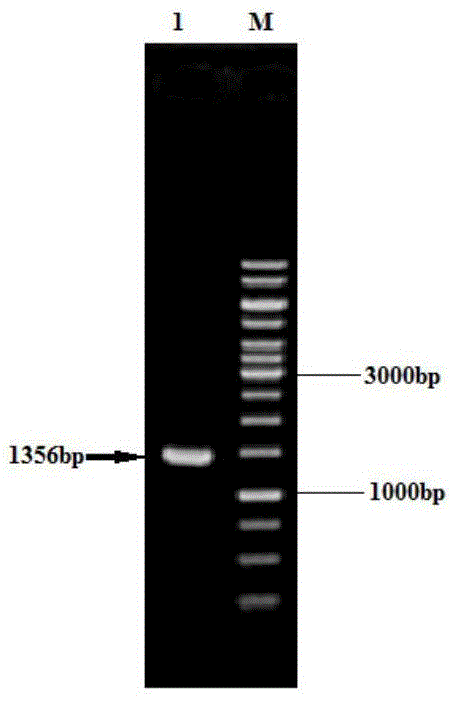
[0065] 4) using the PCR method to amplify serA with the genomic DNA of Corynebacterium glutamicum ATCC13032 as a template Δ197 Gene fragment.
[0066] 5) The purified PCR product was double-digested with BamHI and EcoRI and connected to the expression vector pXMJ19-p...
Embodiment 3
[0080] The codon optimization of embodiment 3pssA gene
[0081] In order to further increase the expression of pssA gene in C. glutamicum, the codon optimization of pssA gene was carried out. The relevant design principles are as follows: (1) Analyze the frequency of degenerate codons in the pssA gene and C. glutamicum, and select the most preferred codons first, and some secondary preferred codons can also be used; (2) The amino acid sequence of the original pssA gene cannot Change.
[0082] Comparison of the nucleotide sequence between the optimized pssAM and the original pssA, the codon optimization of the pssA gene has changed a total of 275 bases, accounting for 20.3% of the total bases of the pssA gene, and the expression of PS in Corynebacterium glutamicum has increased more than 15%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com