Method for identifying rice DNA identities and application thereof
A rice and identity technology, applied in special data processing applications, instruments, electronic digital data processing, etc., can solve the problems of the small number of SSR markers and the inability to fully reflect the genetic background of breeding materials, etc., and achieve simple operability, reliability and reliability Operability, the effect of eliminating statistical errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Embodiment 1 Rice Variety Standard Gene Fingerprint Detection Parameter Determination
[0076] 1. Number of SNP markers
[0077] On the one hand, the more the number of SNP markers, the higher the resolution of DNA differences between varieties; on the other hand, the more the number of SNP markers, the higher the cost of detection and analysis.
[0078] In order to determine the optimal number of markers that can meet the identification of DNA differences between varieties, in this embodiment, the rice genome-wide breeding chip Rice60K detection data from 195 micro-core germplasm resources in the world were used to analyze (Chen et al., Ahigh- density SNP genotyping array for rice biology and molecular breeding. Mol Plant. 2014, 7: 541-553).
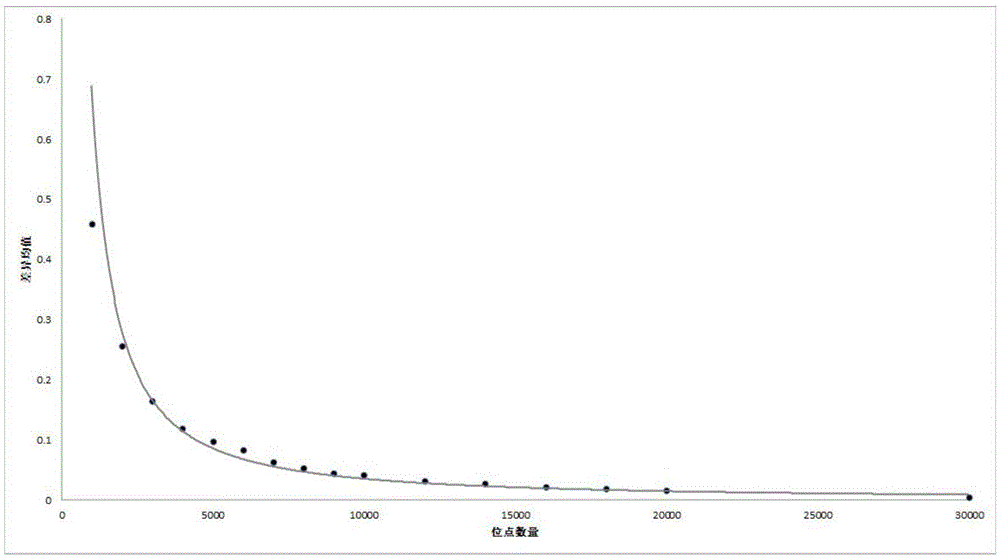
[0079] There are about 44,000 high-quality SNP sites on the rice genome-wide breeding chip Rice60K chip, from which 1,000 to 30,000 sites are randomly sampled for detection, and the genetic distance between two varieties is calc...
Embodiment 2
[0106] Example 2 Using Rice6K Genome-wide Breeding Chip to Construct Standard Gene Fingerprint Database of Rice Varieties and Perform DNA Identification
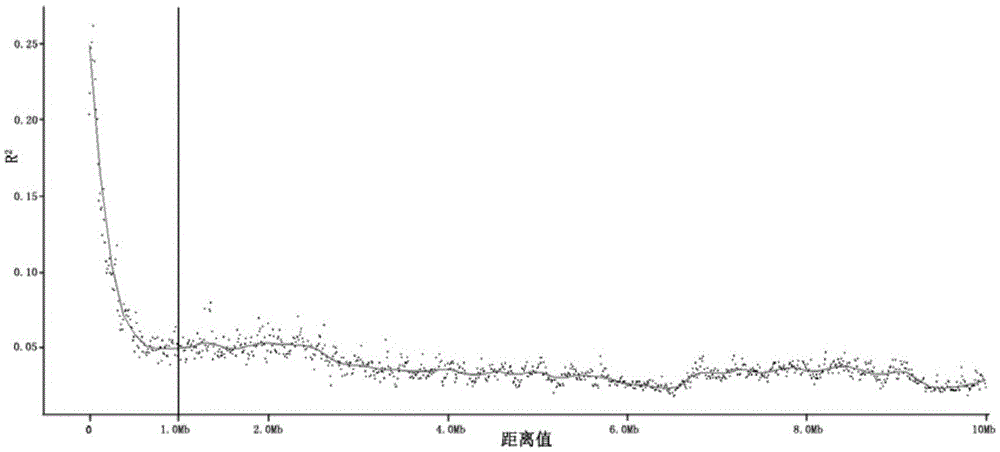
[0107] The markers on the rice genome-wide breeding chip Rice6K are selected from the resequencing data of 520 germplasm resources, representing genetic diversity, a set of markers evenly distributed on the 12 chromosomes of rice, and the number of high-quality markers is about 4500 .
[0108] The chip is the InfiniumSNP chip platform of Illumina Company. The biggest feature of this platform is that the results are stable and reliable. For the high-quality loci that have been tested and selected, the accuracy rate is 99.9% (Yu et al., Awhole-genomeSNParray (Rice6K) forgenomicbreedinginrice.PlantBiotechnolJ.2014 , 12:28-37). According to the detection parameters determined in Example 1, the breeding chip fully meets the conditions for detecting standard gene fingerprints.
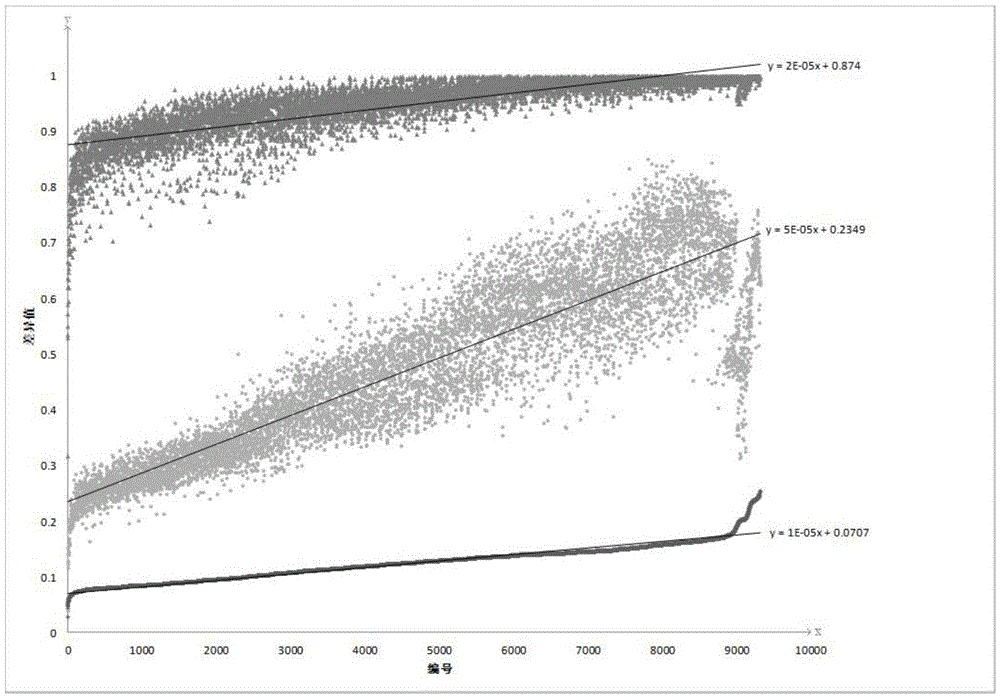
[0109] In this embodiment, the construction of a r...
Embodiment 3
[0152] Example 3 Identification of Rice Variety "Longjing 31" Using Standard Gene Fingerprints
[0153] 1. Material Description
[0154] The conventional japonica rice "Konyu 131" is native to Japan. In 1990, it was introduced and bred by the Rice Research Institute of Heilongjiang Academy of Agricultural Sciences from Jilin Academy of Agricultural Sciences.
[0155] Another conventional japonica rice "Longjing 31" was obtained by using "Longhua 96-1513" as the female parent and "Kendao 8" as the male parent, inoculated with its F1 anthers for in vitro culture, and then selected by pedigree method. In 2011, it passed the examination and approval of Heilongjiang.
[0156]The performance of variety approval: Compared with the control "Kongyu 131", the yield and rice blast resistance of "Longjing 31" were significantly enhanced, and other phenotypic traits such as plant and leaf morphology were significantly different from the control. According to the relevant regulations on ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com