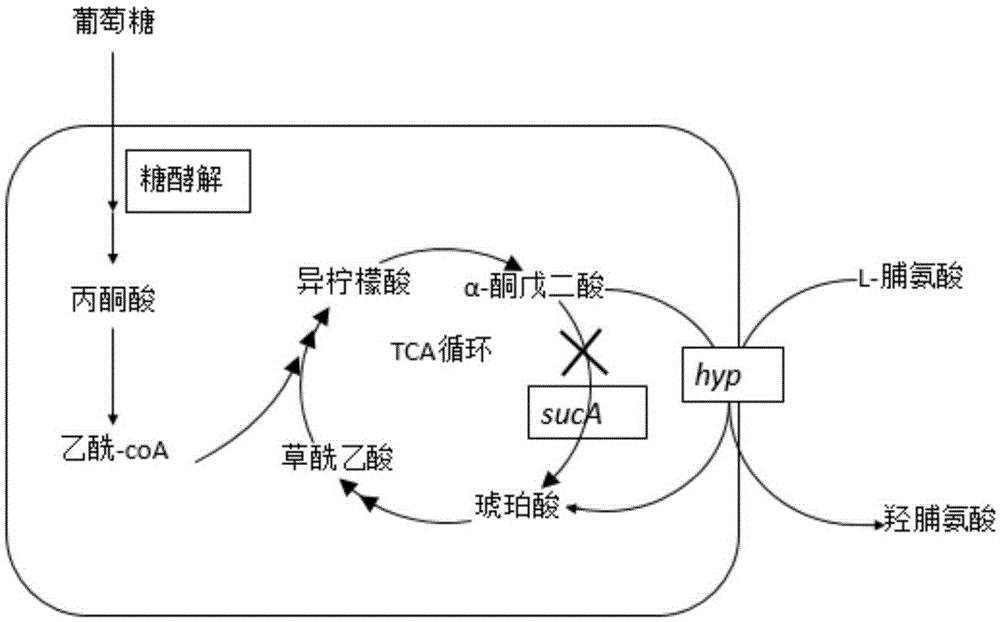
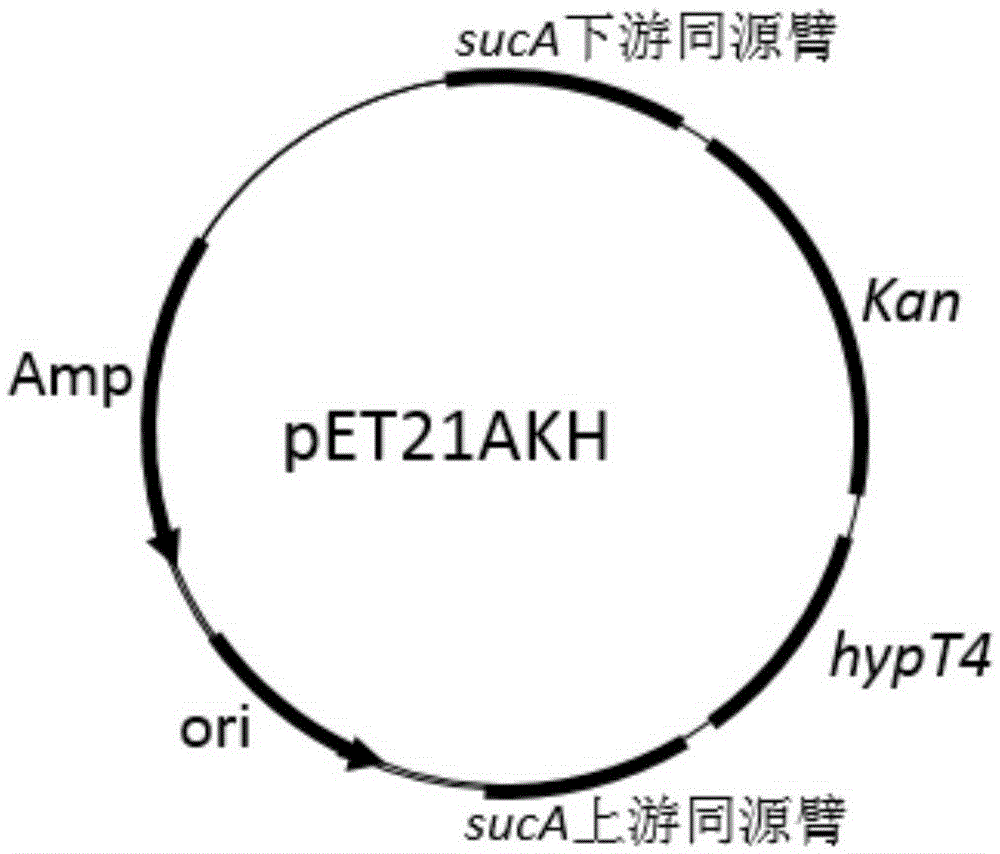
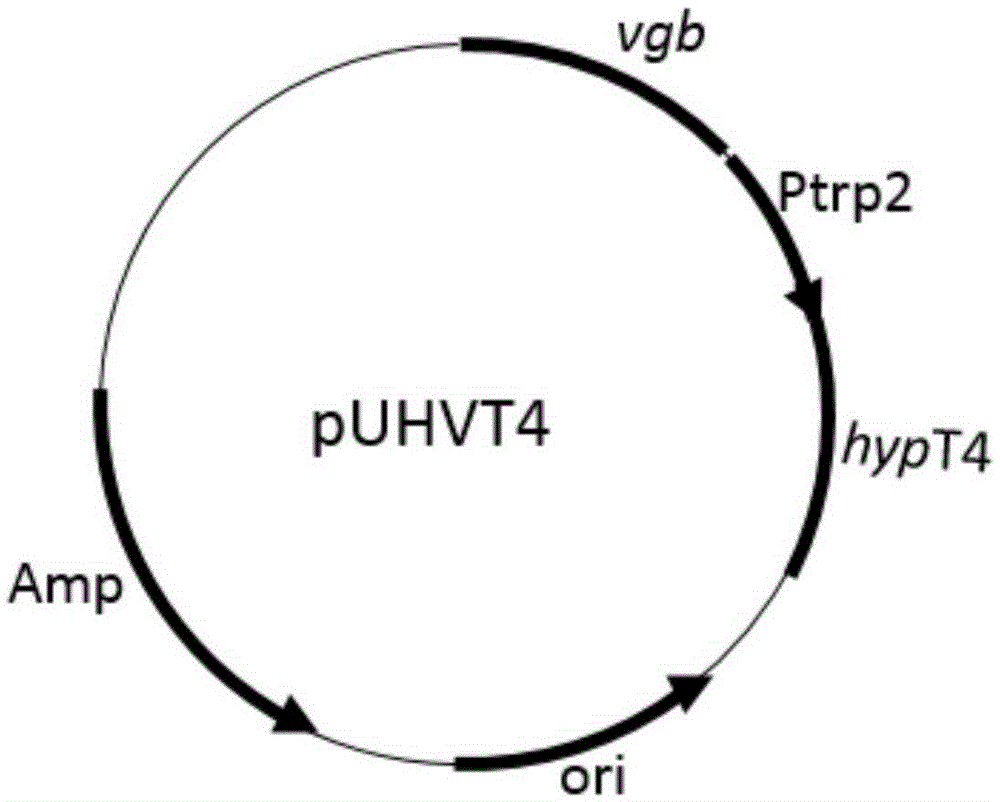
Construction of high-yield trans-4-hydroxyproline sucA gene knockout bacteria
A proline and trans technology, applied in the field of high-yield trans-4-hydroxyproline strain construction, can solve the problems of complexity, high cost, and waste
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Construction of vectors containing targeting fragments
[0026] Using the genome of Escherichia coli as a template, the upstream and downstream source arms of the sucA gene were obtained by PCR; using the plasmid pKD4 as a template, the Kan resistance gene was obtained by PCR; then the three were added at a molar ratio of 1:1:1 for fusion PCR to obtain a preliminary targeting fragment containing the upstream and downstream homology arms of the sucA gene and the Kan resistance gene (when designing primers, the 5' end of the upstream homology arm was introduced into NdeI, the 5' end of the downstream homology arm was introduced into XhoI, and the upstream Restriction sites HindIII and BamHI were introduced between the homology arm and the Kan resistance gene. Ligate the fusion fragment with the vector pUCm-T, screen through the Kana resistance plate, carry out NdeI and XhoI double digestion of the correct plasmid, and connect it with the same double digestion p...
Embodiment 2
[0027] Example 2: sucA gene knockout
[0028] ① Prepare the electroporation competence of Escherichia coli BL21(DE3)ΔputA: transfer the plasmid pKD46 (temperature-sensitive plasmid) into the prepared electroporation competence of Escherichia coli BL21(DE3)ΔputA, and then follow the preparation method of electroporation competence , to prepare the corresponding electroporation competent.
[0029] ② Gene knockout: Electrotransfer the targeting fragment into the prepared electroporation competent, and use the Red recombination system to realize the recombination exchange between the targeting fragment and the genome. The revived strains were spread on the Kanna plate for preliminary screening.
[0030] ③Colony PCR verification: Pick a single colony from the plate and save it on another plate at the same time, put the picked colony into sterile water, follow the normal PCR system, use the verification primer as the primer, and run the gel to verify the size of the PCR product . ...
Embodiment 3
[0033] Example 3: Transformation into plasmid pUHVT4, followed by single-factor optimization of fermentation conditions.
[0034] To prepare Escherichia coli BL21(DE3)ΔputAΔsucA competent for transformation, the plasmid pUHVT4 was transformed into competent cells, the transformation system: plasmid 3 μl, KCM 10 μl, water 37 μl, competent 50 μl. After mixing, ice bath for 30 minutes, heat shock at 42°C for 90s, then place on ice for about 5 minutes, add 600 μl of liquid LB medium, recover at 37°C for about 1 hour, and spread ampicillin plate. Afterwards, a single colony was picked and cultured, the plasmid was extracted, and verified by enzyme digestion. Correct bacterial storage will be verified for subsequent fermentation optimization.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com