Carp spring viremia virus detection kit based on pyrosequencing
A technology of spring carp viremia and pyrosequencing, which can be used in the determination/inspection of microorganisms, biochemical equipment and methods, etc., and can solve problems such as cumbersome operations.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1. Preparation of primers for detection of carp spring viremia virus, viral hemorrhagic sepsis virus and infectious hematopoietic organ necrosis virus
[0073]The material for detecting the fingerprint sequence of carp spring viremia virus (SVCV) includes a sequencing primer SVCV-S for the fingerprint sequence of carp spring viremia virus and a pair of primers SVCV-P for amplifying the fingerprint sequence of carp spring viremia virus. The fingerprint sequence of the carp spring viremia virus is nucleotides 511-526 of SEQ ID No. 1 in the sequence listing. The SVCV-S is the single-stranded DNA shown in SEQIDNo.4 in the sequence listing; the SVCV-P is the single-stranded DNA (SVCV-P-F) shown in SEQIDNo.2 and SEQIDNo.3 in the sequence listing A pair of primers composed of single-stranded DNA (SVCV-P-R), the 5' end of the SVCV-P-R is labeled with biotin, and the amplification product of SVCV-P is the 454th-591st nucleotide of SEQIDNo.1 in the sequence table For the...
Embodiment 2
[0079] The optimization of embodiment 2, SVCV-P, VHSV-P and IHNV-P amplification conditions
[0080] 1. Construction of recombinant vector
[0081] Replace the sequence between the BamHI and HindIII recognition sites of the pMD18-Tsimple vector with the nucleotide sequence shown in SEQIDNo. The vector was named PMD18-T-SVCV.
[0082] Replace the sequence between the HindIII and XhoI recognition sites of the pMD18-Tsimple vector with the nucleotide sequence shown in SEQIDNo. The vector was named PMD18-T-VHSV.
[0083] Replace the sequence between the HindIII and BamHI recognition sites of the pMD18-Tsimple vector with the nucleotide sequence shown in SEQIDNo. The vector was named PMD18-T-IHNV.
[0084] 2. Optimization of SVCV-P amplification conditions
[0085] The amplification conditions of SVCV-P were optimized with the following PCR amplification system: 2 μL of PMD18-T-SVCV vector at a concentration of 50 ng / μl, 10 μL of 5×PCR buffer, 0.25 μL of 5 U / μl Taq hot-start e...
Embodiment 3
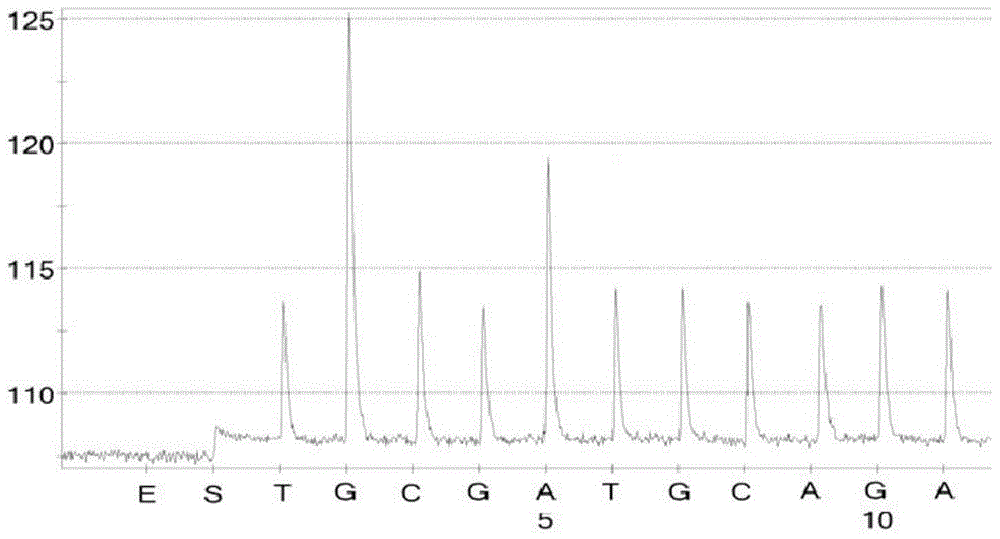
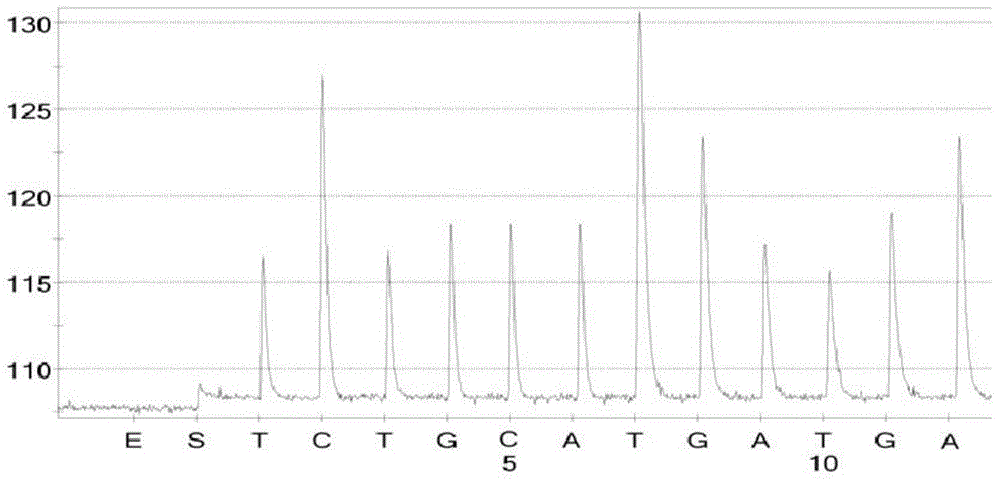
[0093] Embodiment 3, identification of SVCV, VHSV and IHNV by pyrosequencing
[0094] The RNAs of SVCV, VHSV and IHNV were extracted to obtain SVCV total RNA, VHSV total RNA and IHNV total RNA at a concentration of 40 ng / μL, respectively.
[0095] 1. Identification of SVCV by pyrosequencing
[0096] 1.1 PCR amplification
[0097] 50 μL PCR amplification system: 2 μL of SVCV total RNA at a concentration of 40 ng / μL, 10 μL of 5×PCR buffer, 0.25 μL of 5 U / μL Taq hot start enzyme, 0.5 μL of SVCV-P-F at a concentration of 10 pmol / μL, and SVCV-P-F at a concentration of 10 pmol / μL P-R 0.5 μL, make up the volume to 50 μL with DEPC water.
[0098] The amplification conditions were: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s, annealing at 58°C for 30 s, extension at 72°C for 30 s, and 50 cycles; extension at 72°C for 10 min, and ending at 4°C.
[0099] The obtained PCR products were subjected to agarose gel electrophoresis ( figure 1 ), the results showed that...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com