Cervus nippon PRDX4 gene, cloning method thereof and encoding protein
A cloning method and gene coding technology, which can be applied in the field of genetic engineering and can solve problems such as difficulty in the sequence of sika deer PRDX4 cDNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] The extraction (SunH etc., 2012) of embodiment 1 staghorn periosteal cells and the extraction of mRNA
[0030] After the periosteum of the sika antler peduncle obtained from the experimental deer farm of the Chinese Institute of Special Products of Agricultural Sciences was retrieved, according to SunH et al. (SunH, YangF, ChuW, ZhaoH, McMahonC, LiC. : e47367) method to process the obtained periosteum, and obtain the periosteum sensitized zone cells of the sika deer.
[0031] Place the cells in the sensitized area of the antler horn periosteum on the 75cm of the DMEM medium containing 20ml of 10% fetal bovine serum and 1% penicillin and streptomycin 2 Culture flask, 37°C, 5.00% CO 2 Cultivate for 2 days under the same conditions; discard the old culture medium, replace with 20ml of the same new culture medium as above and continue to culture for 1 day under the same conditions to obtain cells in the sensitized area of the periosteum of Stalk antler.
[0032] The c...
Embodiment 2
[0033] Example 2 Primer Design
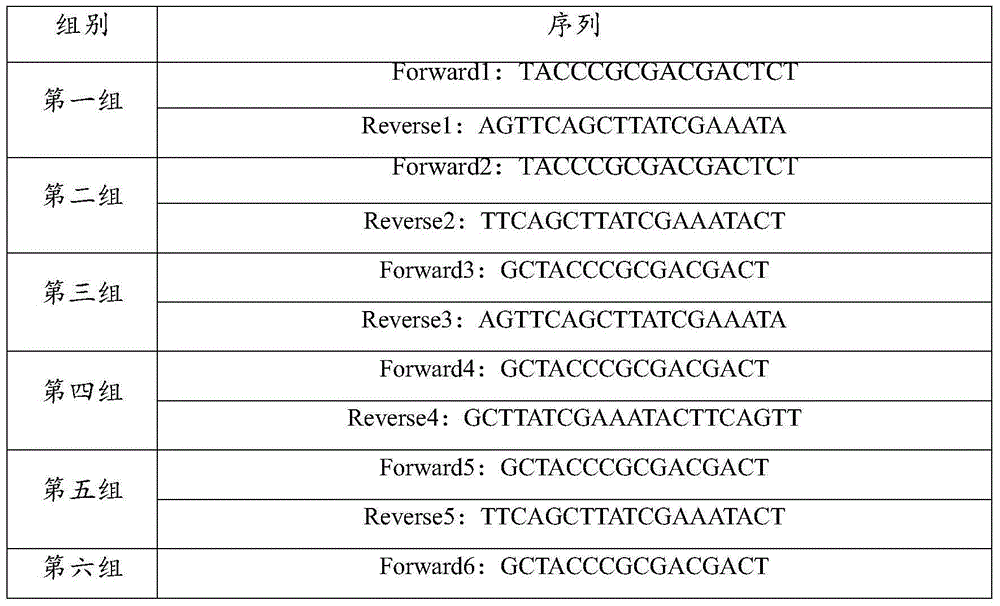
[0034] Primers were designed according to the bovine PRDX4 mRNA sequence, and 11 sets of primers were designed, as shown in Table 1. Amplification was performed using these 11 sets of primers.
[0035] Table 111 sets of primer sequences
[0036]
[0037]
[0038] The primers from the first group to the ninth group were all primers designed by Primer software, and the target product was not obtained by using the nine sets of primers for amplification. The tenth set of primers is two sequences cut directly from the two ends of the bovine PRDX4 mRNA sequence as primers, and no target product is obtained when using this primer set to amplify. The tenth set of primers is to select two sequences of upstream and downstream primers near the complementary sequence of the deer PRDX4 mRNA start codon and stop codon, and after comprehensive evaluation of the CG content, annealing temperature, dimer and complementarity of the primers, and then A sm...
Embodiment 3
[0040] Embodiment 3 bioinformatics analysis
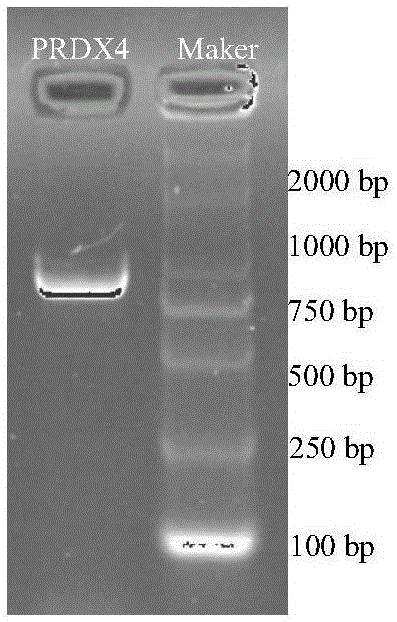
[0041]The sequencing results showed that the mRNA sequence of deer PRDX4 was cloned for the first time in this experiment. Using the cDNA of the cells in the periosteum sensitized region of Antlers antlers as a template, and using the sequences shown in SEQIDNO.3-SEQIDNO.4 as primers, PCR amplification was performed, and the amplification results were detected by nucleic acid electrophoresis. The results are as follows: figure 1 shown. from figure 1 It can be seen that there is a bright single band in the nucleic acid electrophoresis lane with a size of about 800-900bp, the size of the PRDX4 fragment amplified by PCR is basically the same as that of other species, and the actual length of the cloned mRNA sequence obtained is 819bp. Through bioinformatics analysis, it was found that some physical and chemical properties and biological processes of deer PRDX4 protein were basically similar to those of cattle, humans and other speci...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com