A detection method and application of degraded dna from spoilage material
A detection method and technology for testing materials, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as allele loss and low detection rate of loci, and achieve accurate screening results and high forensic application value. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 Acquisition of DNA genetic markers located in the nucleosome core region
[0026] Its DNA genetic markers include SNVs (single nucleotide variation sites), Indels (insertion-deletion polymorphism sites), and STRs (short tandem repeat polymorphism sites).
[0027] experimental method:
[0028] (1) Collect white blood cells from the peripheral blood of healthy people, use micrococcal nuclease (micrococcal nuclease, MNase) to digest the white blood cells after blood lysis; add CaCl at a final concentration of 1mM 2 The solution and micrococcal nuclease with a final concentration of 3U / ul were incubated at 37°C for 3 hours to release the nucleosome core, and the DNA in the digested white blood cells was extracted using the phenol-chloroform method to obtain the nucleosome core in human blood tissue Region DNA;
[0029] (2) Use the Illumina Hiseq 2000 Genome Analyzer analysis platform to sequence the obtained nucleosome core region DNA;
[0030] (3) Use bwa (0.6...
Embodiment 2
[0040] Example 2 Designing primers by taking partial labeling of the obtained nucleosome core region as an example
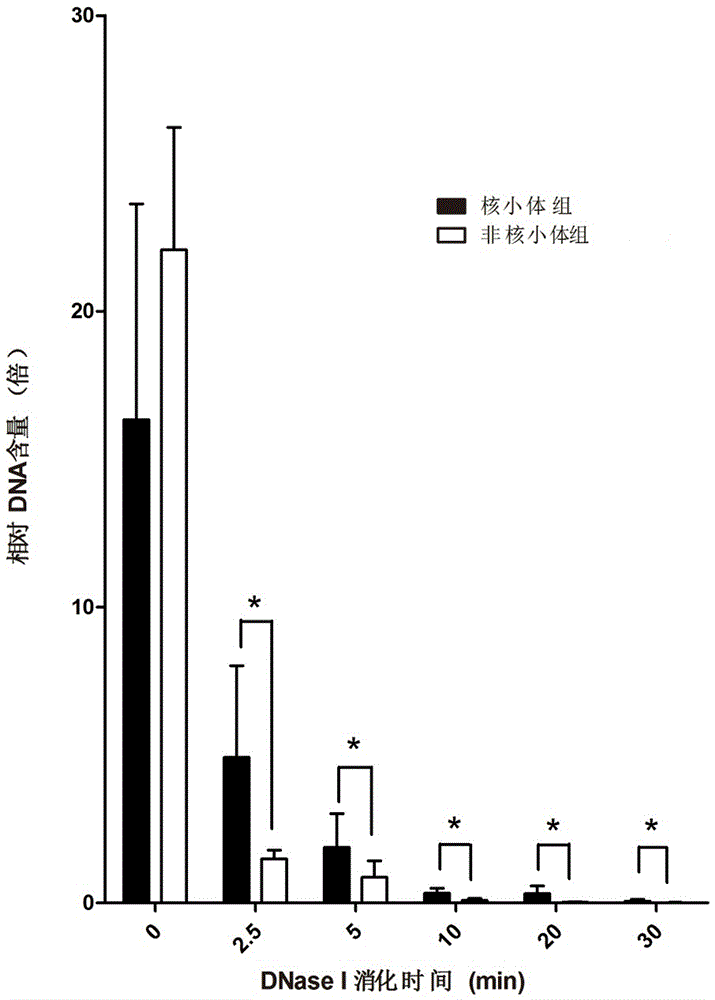
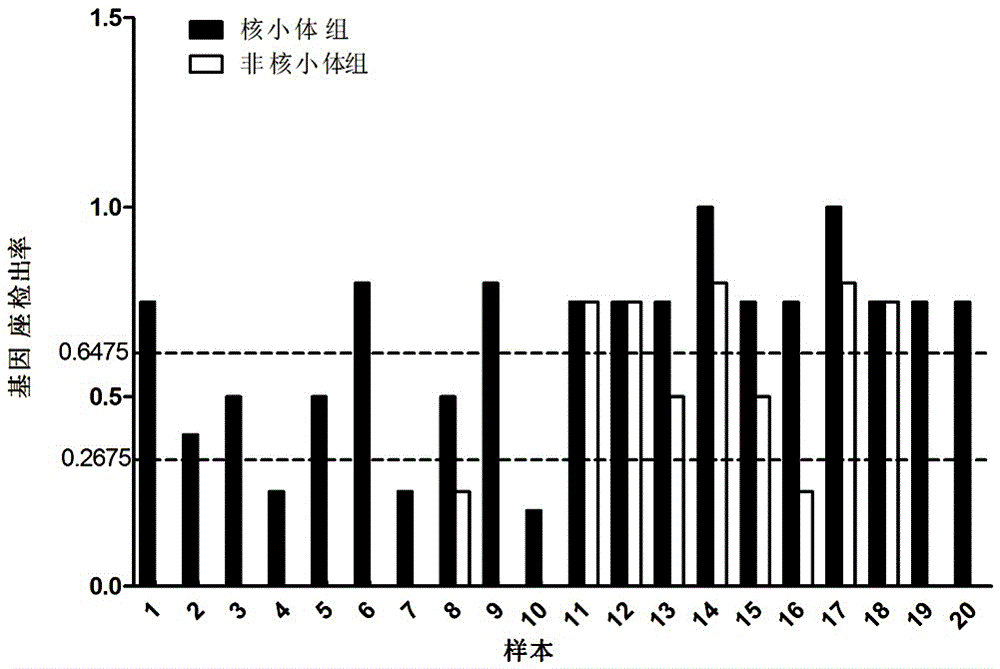
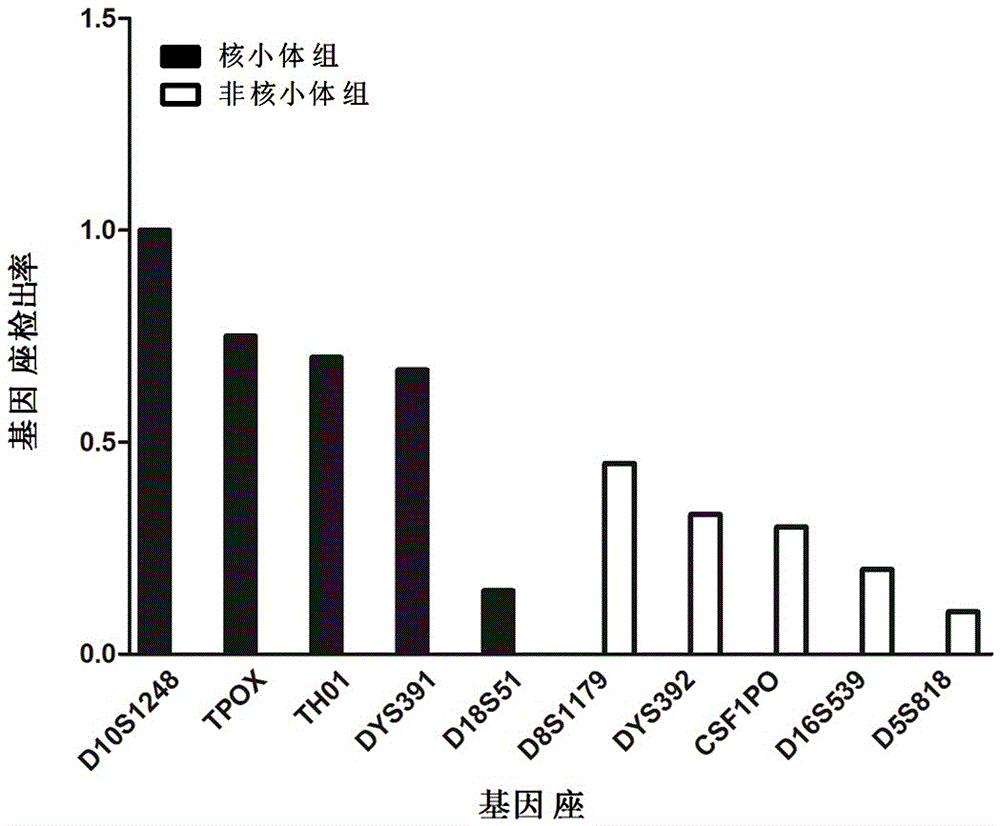
[0041] The STRs marker data obtained in Example 1 were compared with 44 commonly used genetic markers in forensic science. There were 5 STR loci successfully matched, and these 5 STR loci were selected as nucleosome group STRs, namely D10S1248, D18S51, TH01, TPOX and DYS391, see the markers marked with two asterisks in Table 3. Five STR loci were randomly selected from the remaining 39 loci on the unalignment as non-nucleosome group STRs, namely CSF1PO, D5S818, D8S1179, D16S539 and DYS392, see the marker marked with an asterisk in Table 3. Details of selected genetic markers commonly used in forensic science located in different regions of nucleosomes are given in Table 3. By redesigning the primers, the lengths of the amplified fragments of the above 10 selected STRs are all less than the length of the nucleosome core fragment of 147bp. The information of the ...
Embodiment 3
[0048] Example 3 Manual preparation of degraded samples and extraction of degraded DNA
[0049] The blood samples of 5 different individuals (A, B, C, D and E) were taken as samples, and the DNA in the blood was extracted respectively (using QIAamp DNA Blood Midi kit, Qiagen, Germany), and 10 µg of DNA was taken at a concentration of 0.01 U / μl of the digestive enzyme DNase I for digestion, and after digestion to 0, 2.5 min, 5 min, 10 min, 20 min and 30 min each time point, samples were taken, and 30 samples of artificially degraded samples were obtained.
PUM
| Property | Measurement | Unit |
|---|---|---|
| correlation coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com