Method for detecting staphylococcus aureus and enterotoxin A gene in food
A Staphylococcus aureus technology, applied in the detection of target genes, primer pairs and probes, can solve the problems of limited detection of Staphylococcus aureus, false negatives, false positives, etc., to achieve practical, specific, and results. Determining simple effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Establishment of PMA-qPCR detection method
[0056] Step 1, design of detection primers and related probes
[0057] Through comparative genomics analysis, the specific detection target gene RpiR of Staphylococcus aureus was obtained. Download all the nucleic acid sequences of the RpiR gene from NCBI GenBank, and perform homology comparison analysis to find the conserved region of the RpiR gene. The base sequence information of the target sequence is shown in SEQ ID NO:1. At the same time, the nucleotide sequence of the Staphylococcus aureus enterotoxin A gene (sea) was downloaded from GenBank, and then homologous comparison analysis was performed to find the conserved region of the gene. The base sequence information of the enterotoxin A gene is shown in SEQ ID NO:2.
[0058] Input the base sequences of RpiR and sea into Beacon designer7.0 respectively, design primer pairs and probes in the conserved regions of the two genes, set the GC% range to 40-60%, and the prod...
Embodiment 2
[0076] Performance Evaluation of PMA-qPCR Assay
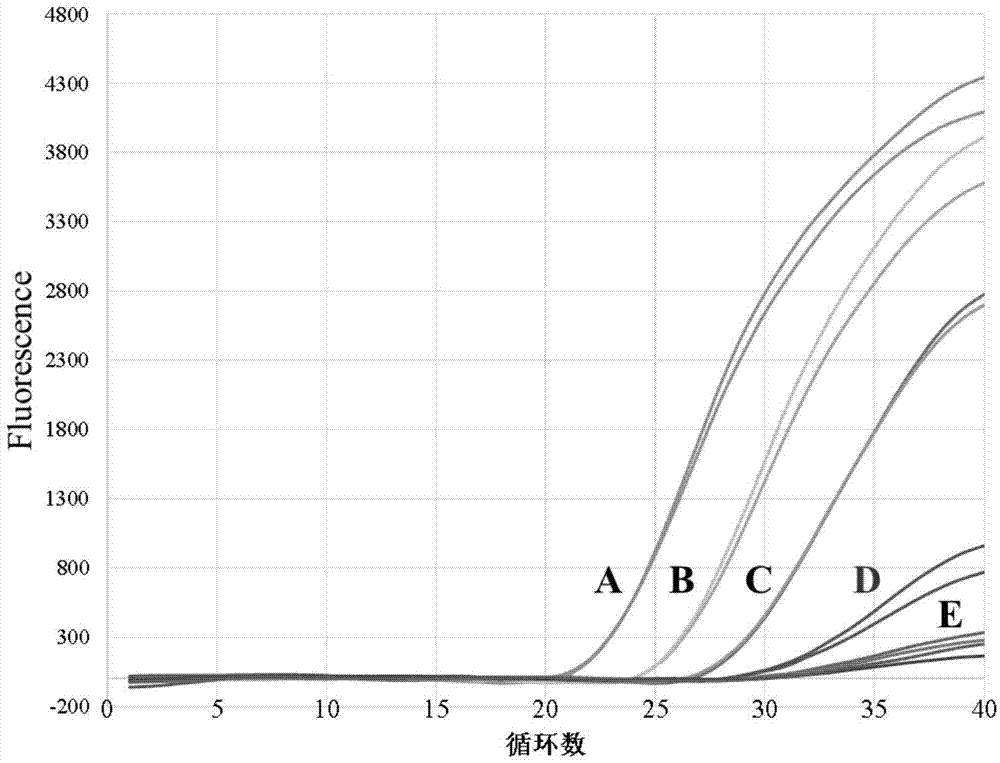
[0077] The Staphylococcus aureus standard strain ATCC13565 (sea+) and the standard strain ATCC25923 (sea-) cultured overnight were serially diluted after plate counting. Weigh 25g of quick-frozen pork dumplings negative for Staphylococcus aureus, add 225mL of normal saline, beat with a homogenizer for 100s, take 900μL of homogenized solution, add 100μL of diluted pure culture, and make 3 parallels for each concentration gradient. Contaminate food samples with standard strains ATCC 13565 and ATCC 25923 alone, so that the concentration of the strains that contaminate the food is about 10 2 cfu / g, 10 3 cfu / g, 10 5 cfu / g and 10 7 cfu / g and mixed strains contaminated food samples (choose two concentration gradients, approximately: 10 7 cfu / g ATCC25923 and 10 3 cfu / g ATCC 13565, 10 7 cfu / g ATCC 25923 and 10 5 cfu / g ATCC 13565, 3 parallels per concentration gradient). Then the food samples were treated with PMA, and the genomi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com