Corynebacterium-glutamicum recombinant strain for increasing conversion rate of L-phenylalanine saccharic acid
A technology of Corynebacterium glutamicum and glutamic acid bar, which is applied in the field of metabolic engineering and can solve problems such as Escherichia coli restriction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
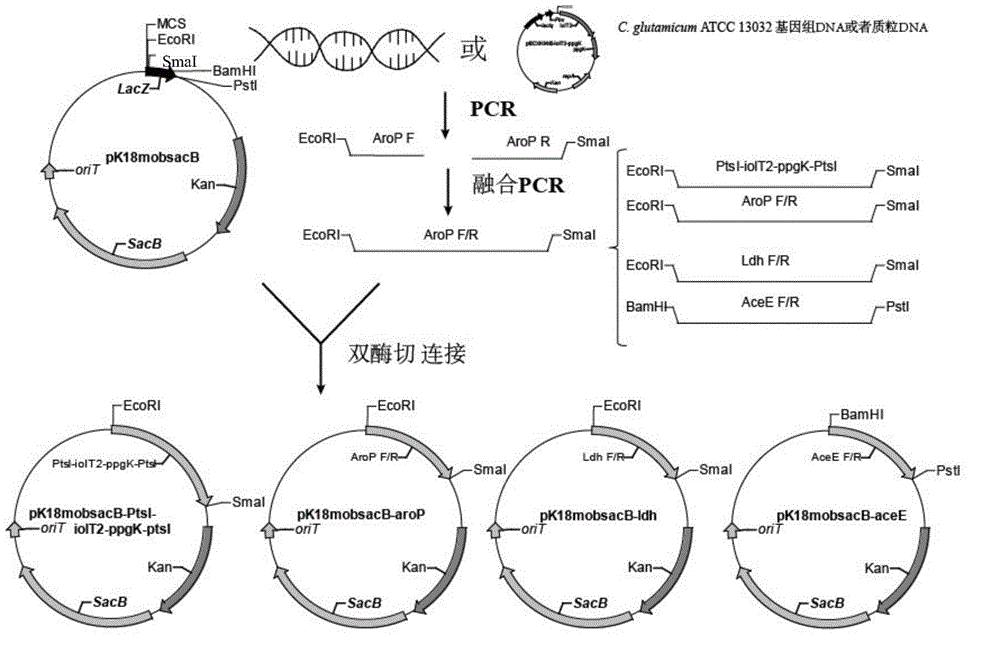
[0060] Example 1 Construction and Verification of pK18mobSacB-ptsI-iolT2-ppgk-ptsI Vector
[0061] The knockout vector pK18mobSacB-ptsI-iolT2-ppgk-ptsI was constructed by two fusion PCRs.
[0062] (1) First three homologous arms: use primers 1# (sequence shown in SEQ ID NO.1) and 2# (sequence shown in SEQ ID NO.2) to amplify to C. glutamicum ATCC 13032 as a template 812bp PtsI upstream homology arm (the downstream has the complementary sequence of the iolT2-ppgK gene cluster, and the upstream primer has an EcoRI restriction site), using primers 3# (sequence shown in SEQ ID NO.3) and 4# (sequence shown in SEQ ID NO.4) with the carrier pECXK99E-iolT2-ppgK as the template amplified to the iolT2-ppgK gene cluster of 2.3Kb size (the upstream has a sequence complementary to the downstream of the PtsI upstream homology arm, the downstream band There is a sequence complementary to the upstream of the PtsI downstream homology arm); with primers 5# (sequence shown in SEQ ID NO.5) and 6...
Embodiment 2
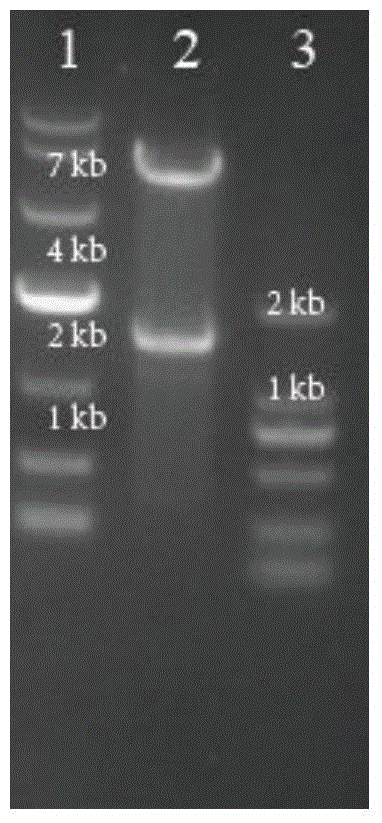
[0067] Example 2 Construction of pK18mobSacB-aroP vector
[0068] With the C.glutamcium ATCC 13032 genome as a template, use primer 9# (sequence shown in SEQ ID NO.9) and primer 10# (sequence shown in SEQ ID NO.10) in Table 2 to amplify to 740bp The upstream homology arm aroP F of the aroP gene is amplified using primer 11# (sequence shown in SEQ ID NO.11) and primer 12# (sequence shown in SEQ ID NO.12) AroP-R-F1 / R1 854bp was deleted in the middle of the downstream homology arm aroP R of the 747bp aroP gene. The fusion PCR method was used to fuse aroP F and aroP R into 1487bp aroP F / R. After the fragment aroP F / R and plasmid pK18mobSacB were digested with EcoRI and SmaI, the aroP F / R was connected to the plasmid vector pK18mobSacB using T4 ligase, transformed into E. coli E.coli JM109, and the plasmid was extracted using restriction enzymes EcoRI and SmaI digestion verification such as image 3 As shown, lane 1 is DL 10000DNAMarker, lane 2 is pK18mobSacB-aroP digested, lane...
Embodiment 3
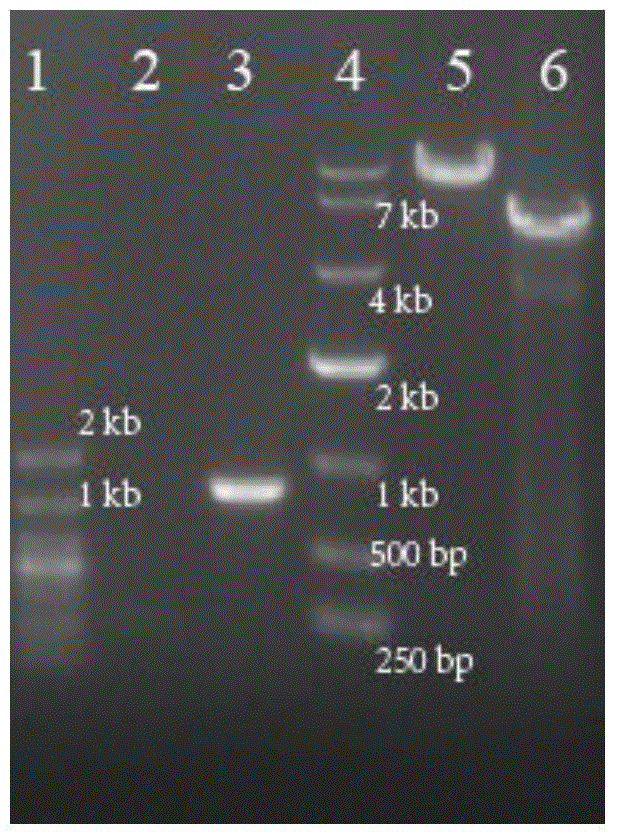
[0069] Example 3C. Validation of glutamicumΔptsI::iolT2-ppgKΔaroP strain
[0070] In Corynebacterium glutamicum C. glutamicum ATCC 13032, the vector pK18mobSacB-ptsI-iolT2-ppgk-ptsI gene knockout constructed by constructing the knock-in iolT2-ppgK gene cluster was transferred into the recombinant strains, and the genome was extracted as shown in Table 2 Primer 7# (sequence as shown in SEQ ID NO.7) and primer 8# (sequence as shown in SEQ ID NO.8) carry out PCR verification (knockout verification primer is inside knockout ptsI gene, Text-PtsI-F and Text-PtsI-R knockout before 800bp, no PCR product after successful knockout) See the results figure 2As shown, the 800bp PCR band amplified from lane 3 is a negative recombinant, and from lane 2, there is no PCR band, indicating that the obtained strain is a recombinant of the ptsI site knock-in gene cluster iolT2-ppgK, It shows that the obtained strain is C. glutamicumΔptsI::iolT2-ppgK. On the basis of the recombinant strain C.glu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com