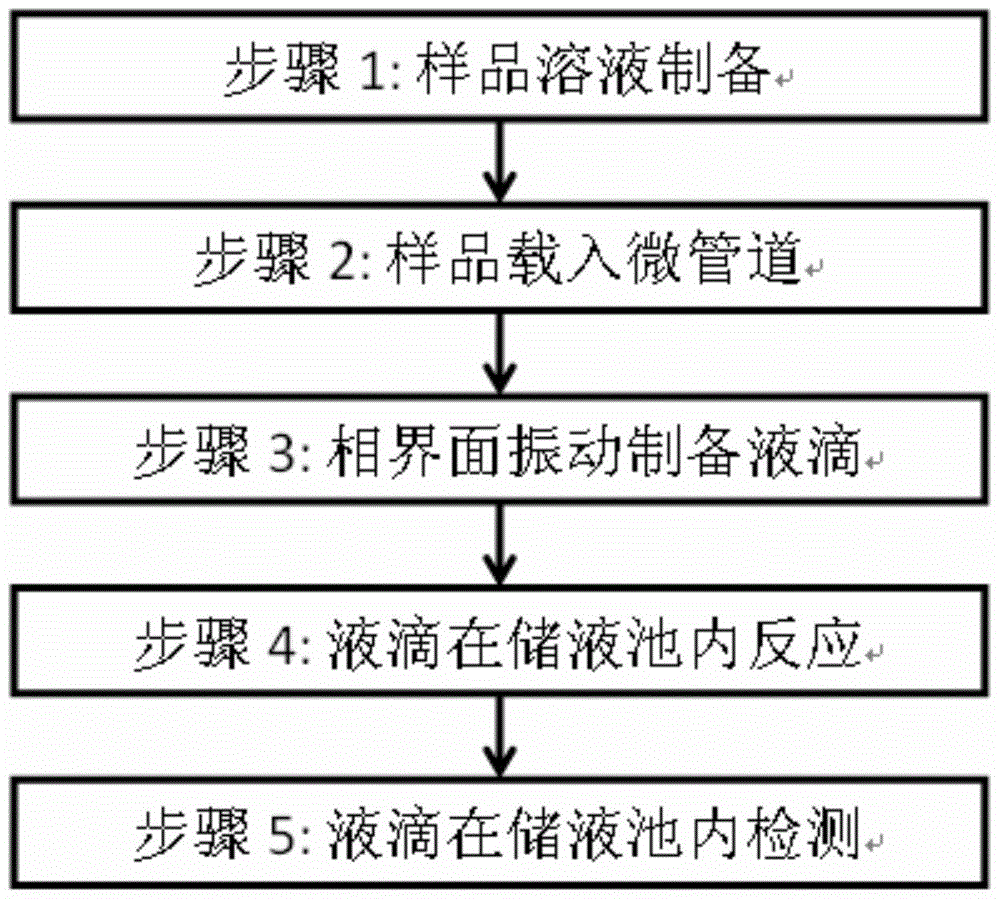
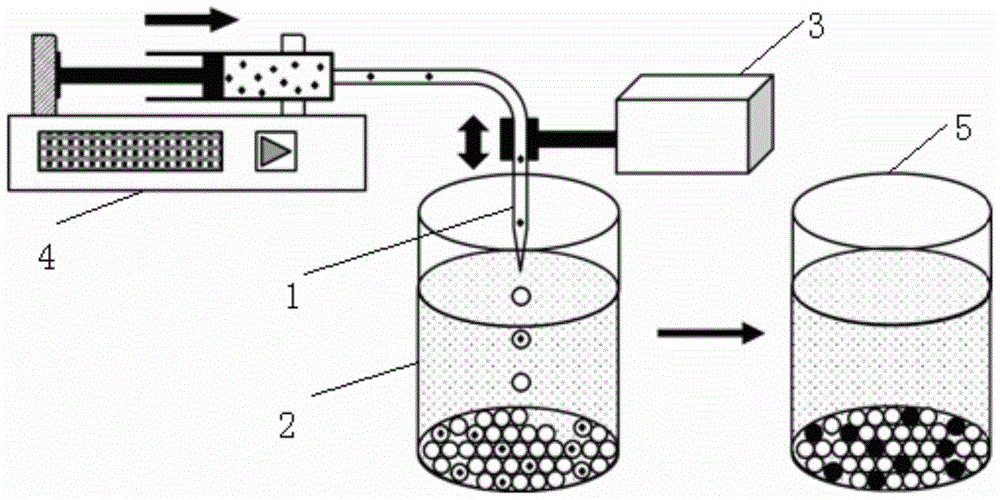
Quantitative analysis method and system for digital nucleic acid amplification based on micro-droplets
A technology for quantitative analysis and nucleic acid amplification reaction, applied in the field of digital nucleic acid amplification quantitative analysis method and system based on micro-droplets, which can solve the problems of high cost and unfavorable digital nucleic acid application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0095] The vibrating device is an electromagnetic dotting timer powered by an AC power supply with a voltage of 6V and a frequency of 50Hz. After the power is turned on, the vibrating piece of the electromagnetic timer vibrates at a frequency of 50 times per second. The amplitude of the vibration is 4mm. The micropipe used is a pointed quartz capillary with a length of 5 cm, an inner diameter of 75 μm, and an outer diameter of 150 μm. The inner diameter of the tip is 30 μm and the outer diameter is 60 μm. One end of the quartz capillary was connected to a 50 μL micro-syringe (Shanghai Gaoge) filled with mineral oil through a Teflon tube with an inner diameter of 300 μm, and the connection between the capillary and the Teflon tube was sealed with epoxy resin. The microsyringe is fixed on a microsyringe pump (Harvard Instruments, USA), and a fixed volume sample is drawn or injected at a set flow rate. The reservoirs used are transparent polystyrene flat-bottomed 96-well plates...
Embodiment 2
[0104] According to the method of Example 1, a droplet with a volume of 4nL was obtained and a nucleic acid amplification reaction was performed; the difference was that the flow rate of the syringe pump was 200nL / s.
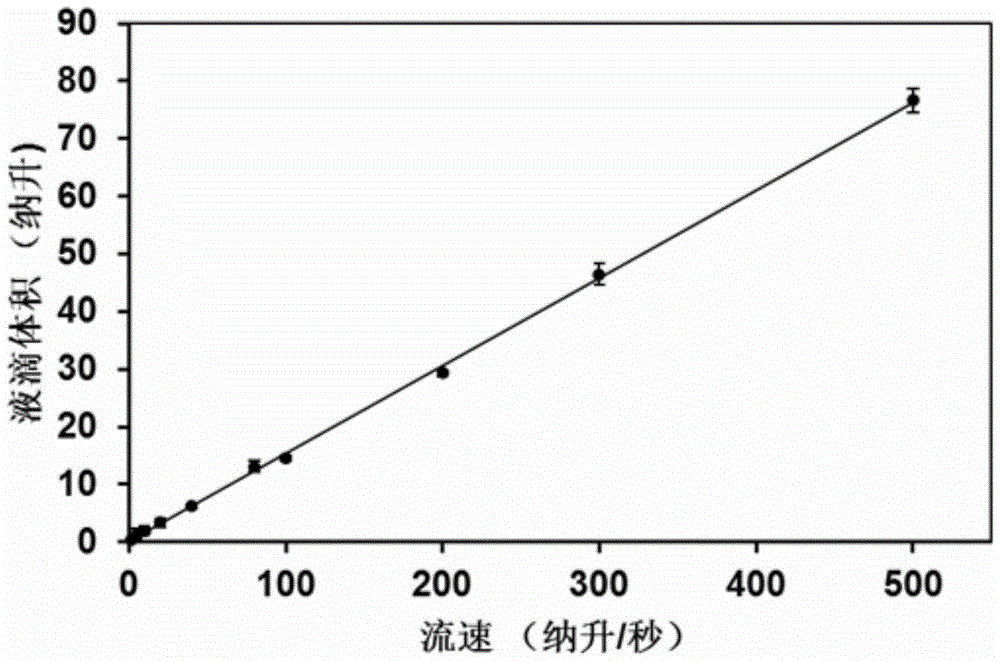
[0105] Generated droplets see Figure 6 , Figure 6 Microscopic view of a droplet generated for Example 2 of the present invention; droplet volume see image 3 . Droplets that produce a fluorescent signal see Figure 7 , Figure 7 It is a schematic diagram of the reaction of droplets with different volumes in Examples 2 to 4 of the present invention.
Embodiment 3
[0107] According to the method of Example 2, a droplet with a volume of 1.7 nL was obtained and a nucleic acid amplification reaction was performed; the difference was that the flow rate of the syringe pump was 80 nL / s, and the injection volume was 2.5 μL.
[0108] Generated droplets see Figure 8 , Figure 8 Microscopic view of a droplet generated for Example 3 of the present invention; droplet volume see image 3 . Droplets that produce a fluorescent signal see Figure 7 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com