Method for knocking out selective marker gene of transgenic pig
A technology for screening marker genes and transgenic pigs, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
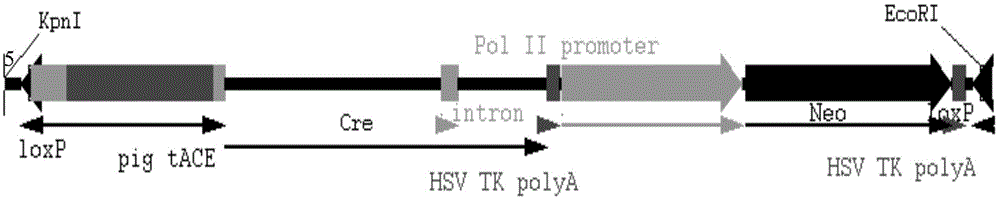
[0028] Example 1 Construction of self-shearing element PCN
[0029] The pig sperm-specific promoter was constructed into the pUC57 cloning vector (provided by the University of Utah, USA) for gene synthesis, named pGENE1 vector (this step was performed by the University of Utah, USA), and double-digested with SalI+AgeI enzymes, the system was 50ul: Carrier 2ul, enzyme 2ul each, 10xbuffer4 plus 5ul, sterile water 31ul, enzyme digestion at 37°C for 2 hours, gel run recovery to obtain a 931bp target fragment, sequenced (nucleotide sequence shown in SEQ ID No.1); and then pACN The carrier (provided by the University of Utah, USA) was double-digested with SalI+AgeI enzyme, and the system was 50ul: 2ul of vector, 2ul of each enzyme, 5ul of 10xbuffer4, 31ul of sterile water, digested at 37°C for 2h, and recovered the target fragment of 5805bp by running the gel ; Ligate the two fragments recovered above, 10ul of the system: 6ul of the recovered small fragment, 2ul of the large fragme...
Embodiment 2
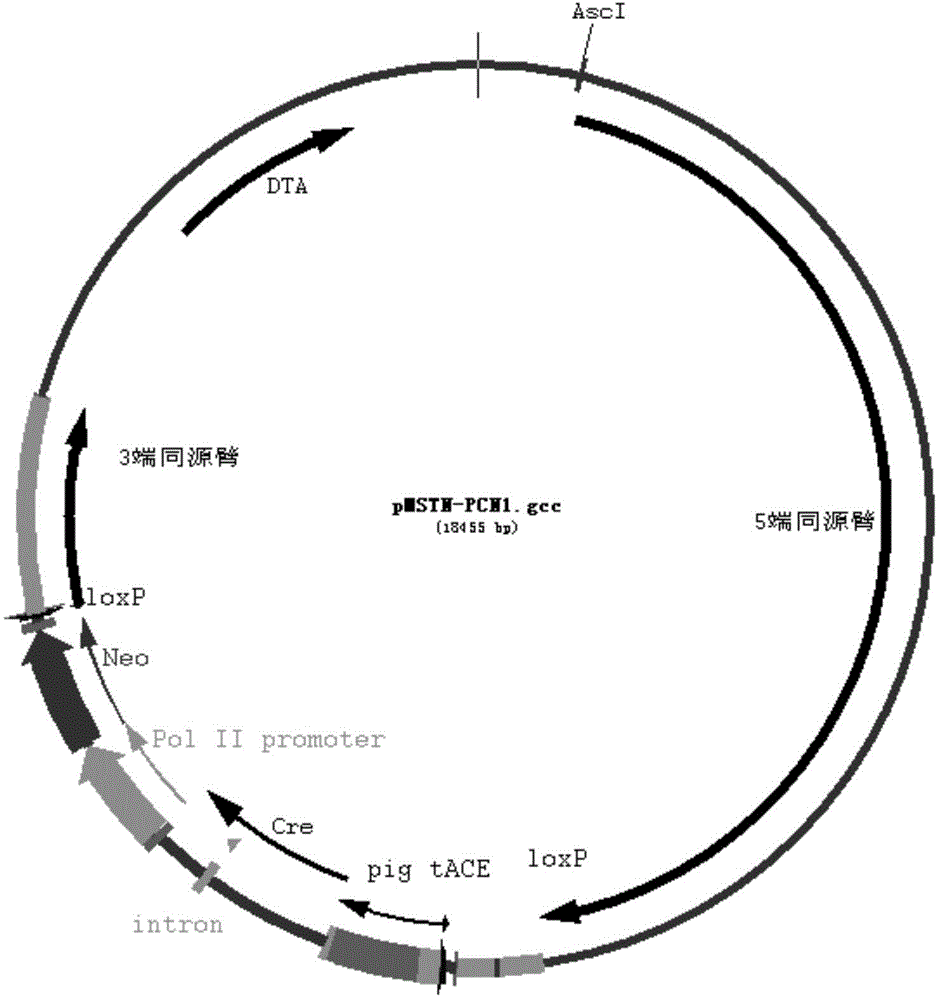
[0030] Example 2 MSTN: construction of marker free targeting vector
[0031] 2. Double-cut the correctly constructed pPCN vector with NheI+NotI, and the system is 50ul: 2ul of vector, 2ul of each enzyme, 5ul of 10xbuffer4, 31ul of sterile water, enzyme digestion at 37°C for 2h, running gel to recover 3620bp Fragment; at the same time, the original pMSTN targeting vector in the laboratory is 50ul with NheI+NotI double enzyme digestion system: 2ul of vector, 2ul of each enzyme, 5ul of 10xbuffer4, 31ul of sterile water, enzyme digestion at 37°C for 2h, running the gel to recover the target 14580 fragments to obtain Backbone fragment, system 10ul: recovered small fragment 6ul, large fragment 2ul, 10xbuffer plus 1ul, T4 ligase 1ul, ligated overnight at 16. The final marker free targeting vector pMSTN-PCN1 vector (nucleotide sequence shown in SEQ ID No.3) for transformation, shaking, small body plasmid, enzyme and positive identification is used for the next step of the experiment (...
Embodiment 3
[0031] 2. Double-cut the correctly constructed pPCN vector with NheI+NotI, and the system is 50ul: 2ul of vector, 2ul of each enzyme, 5ul of 10xbuffer4, 31ul of sterile water, enzyme digestion at 37°C for 2h, running gel to recover 3620bp Fragment; at the same time, the original pMSTN targeting vector in the laboratory is 50ul with NheI+NotI double enzyme digestion system: 2ul of vector, 2ul of each enzyme, 5ul of 10xbuffer4, 31ul of sterile water, enzyme digestion at 37°C for 2h, running the gel to recover the target 14580 fragments to obtain Backbone fragment, system 10ul: recovered small fragment 6ul, large fragment 2ul, 10xbuffer plus 1ul, T4 ligase 1ul, ligated overnight at 16. The final marker free targeting vector pMSTN-PCN1 vector (nucleotide sequence shown in SEQ ID No.3) for transformation, shaking, small body plasmid, enzyme and positive identification is used for the next step of the experiment (see the vector structure for details) figure 2 ). Example 3 Acquisit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com