Characteristic nucleotide sequence, nucleic acid molecular probe, kit and method for identifying branched Cordyceps
A technology of branched Cordyceps and nucleic acid molecules, applied in biochemical equipment and methods, measurement/testing of microorganisms, DNA/RNA fragments, etc., can solve problems such as molecular identification methods of branched Cordyceps that have not yet been found, and achieve a simple and efficient method Short duration and good specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Extraction of Genomic DNA of Cordyceps and Acquisition of ITS rDNA Gene
[0029] Get 20 mg branched Cordyceps samples and place them in a sterile mortar, pour liquid nitrogen into them and grind them quickly, select the DNA extraction kit [Ezup Column Genomic DNA Extraction Reagent from Sangon Bioengineering (Shanghai) Co., Ltd. box (fungi), Cat. No. 13122749Y] to extract DNA from branched Cordyceps. Genomic DNA of Cordyceps sinensis, Cordyceps militaris, Cordyceps guangdong, and cicadae were extracted with reference to the above method.
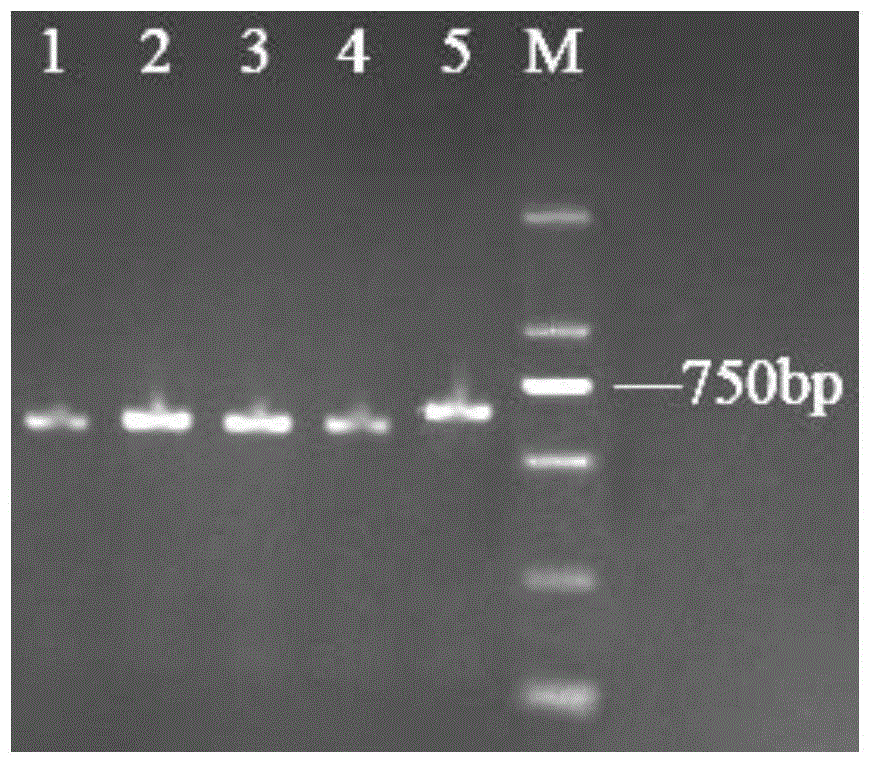
[0030] Take fungal ITS rDNA general primers ITS1 and ITS4, ITS1: 5`-TCCGTAGGTGAACCTGCGG-3`; ITS4: 5`-TCCTCCGCTTATTGATATGC-3`, the primers were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd. The PCR reaction was carried out using the PCR Master Mix kit [Dream Taq Green PCR Master Mix (2×) from Thermo Scientific Company, Cat. No.: 00139170]. The total reaction system is 50.0 μL, PCR Master Mix 20.0 μL, ddH 2 O2 7.0 μL, ITS...
Embodiment 2
[0032] PCR Amplification of Cordyceps Specific Primers
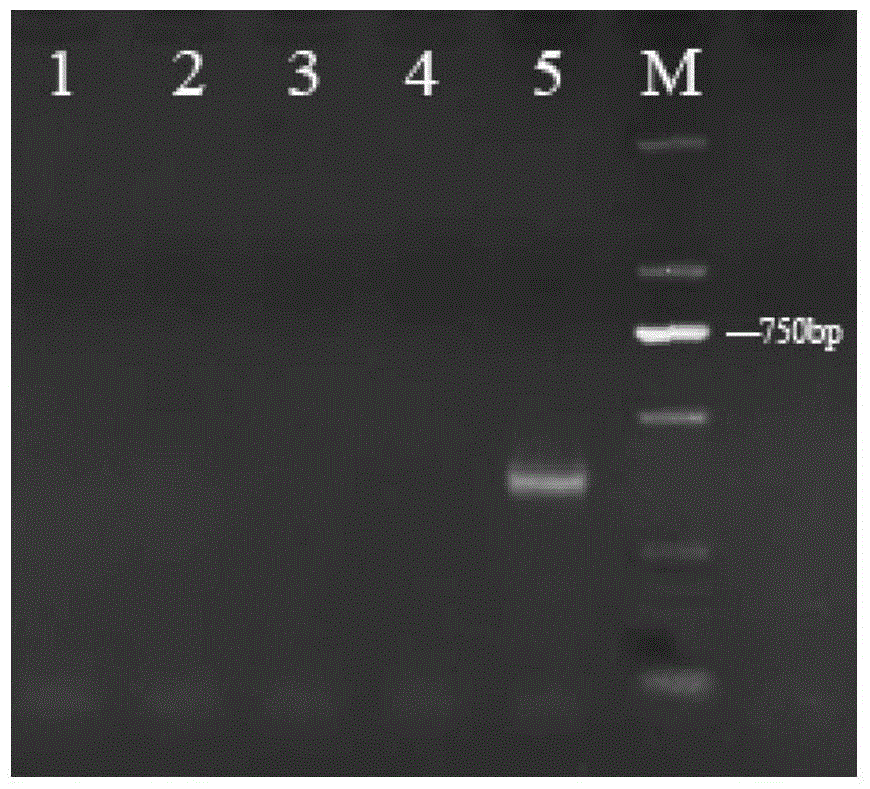
[0033] Specific primers FZF and FZR were designed according to the ribosomal rDNA internal transcription spacers ITS1, ITS2 and the 5.8S rDNA gene sequence between them, FZF: 5`-AGCGGTGGGCGAATGA-3` and FZR: 5`-ACTTTAGCTGCGGGCG- 3`. Primers were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd. Using the ITS rDNA sequence genes of different Cordyceps samples as templates, the total reaction system was 50.0 μL, PCR Master Mix 20.0 μL, ddH 2 O2 7.0 μL, FZF (10 μmol / L) 1.0 μL, FZR (10 μmol / L) 1.0 μL, template DNA (ITS rDNA gene of Cordyceps sinensis, Cordyceps militaris, Cantonese caterpillar fungus, cicadae, branch Cordyceps) 1.0 μL. The reaction program was pre-denaturation at 94°C for 3 minutes, 35 cycles at 94°C for 30s, 30s at 60°C, 30s at 72°C, and 8 minutes at 72°C. Electrophoresis detection of PCR results, electrophoresis as shown in image 3 shown, from image 3 It can be seen that only branched Cordyce...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com