A Quasi-targeted Metabolomics Analysis Method Based on Liquid Chromatography/Mass Spectrometry
A technology of metabolomics and analytical methods, applied in analytical materials, scientific instruments, material separation, etc., to achieve the effect of wide detection linear range, good detection repeatability and response linearity, and improved accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Establishment of UHPLC / QQQ MS-DMRM-based Metabolomics Analysis Method for Serum Pseudo-targets
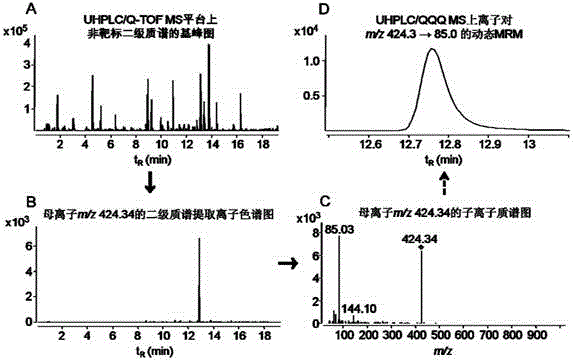
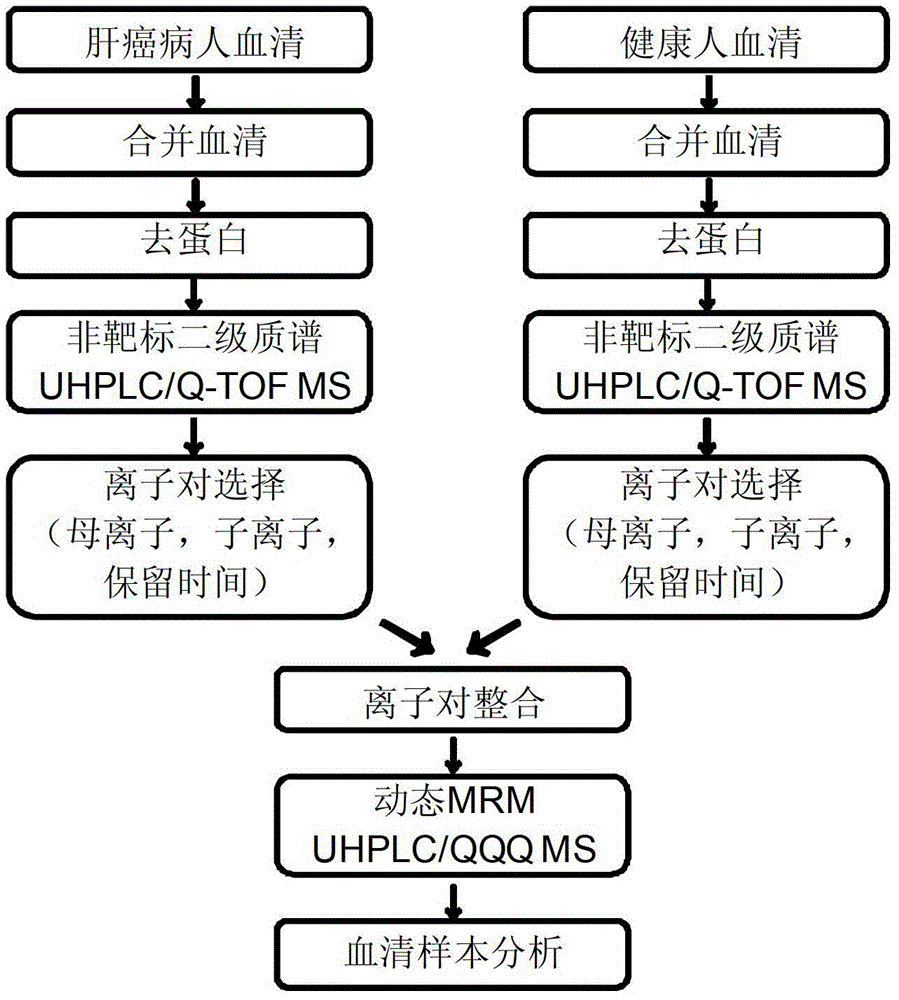
[0026] The process of establishing the metabolomics analysis method based on UHPLC / QQQ MS-DMRM for serum quasi-targets is shown in the attached figure 1 As shown, the specific implementation steps are as follows:
[0027] 1. Combined sample making and preprocessing
[0028] 50 μL of sera from 29 cases of liver cancer patients were pipetted in equal amounts and pooled, and at the same time, 50 μL of sera from 30 normal people were pipetted in equal amounts and combined. Pipette 200 μL of combined serum from liver cancer patients and normal subjects, add 800 μL of acetonitrile to separate proteins, and pipette 800 μL of supernatant to freeze-dry. The freeze-dried product was redissolved in 100 μL pure water for injection analysis.
[0029] 2. Automatic MS / MS data acquisition of metabolites
[0030]Ultra-high performance liquid chromatography (Agilent1200RRLC) / qu...
Embodiment 2
[0041] Example 2: Methodological investigation of metabolomics analysis of serum quasi-targets based on UHPLC / QQQ MS-DMRM
[0042] Mix equal volumes of the pooled serum of normal people and the pooled serum of patients with liver cancer to form a quality control (QC) sample, and perform pretreatment according to the following different objects.
[0043] 1. Repeatability inspection of analytical methods
[0044] Pipette 100 μL of QC serum, add 400 μL of acetonitrile to remove protein, and pipette 400 μL of supernatant to lyophilize. The freeze-dried product was redissolved in 100 μL pure water for injection analysis. Repeat processing of 10 QC serum samples was done in parallel.
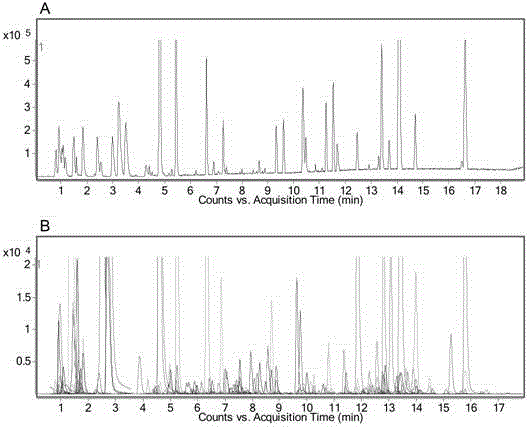
[0045] The 10 QC samples were analyzed by metabolomics on UHPLC / Q-TOF MS and UHPLC / QQQ MS-DMRM platforms respectively. The data measured on the UHPLC / Q-TOF MS platform was matched by XCMS software, and 318 peaks were obtained that were identical to those measured on the UHPLC / QQQMS-DMRM platform. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com