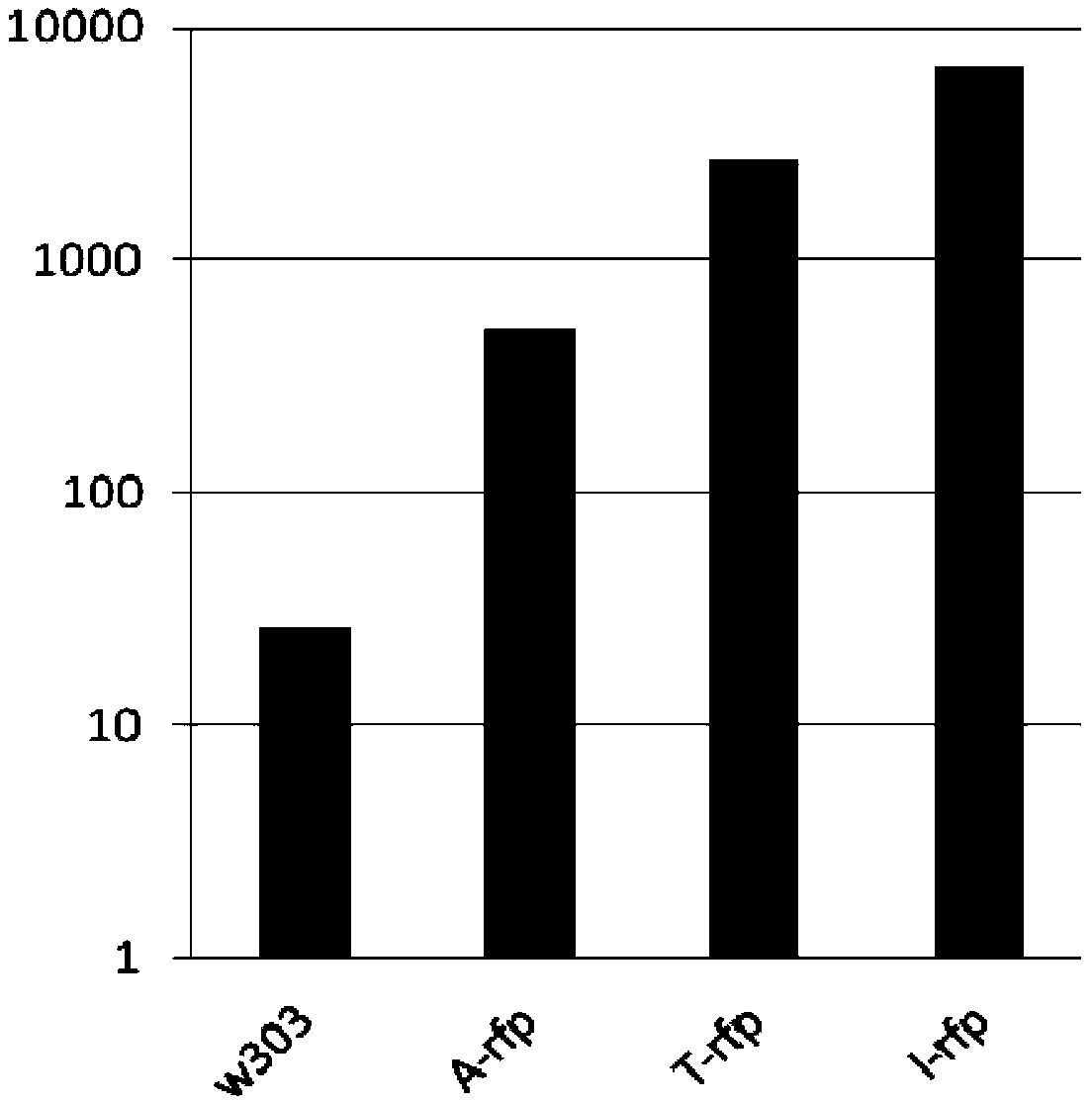Saccharomyces cerevisiae integrated expression vector
A technology of Saccharomyces cerevisiae and chromosome, applied in the field of integrated expression vectors of Saccharomyces cerevisiae, which can solve the problems of increased operation complexity, unstable production performance of engineering bacteria, easy loss of plasmids, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
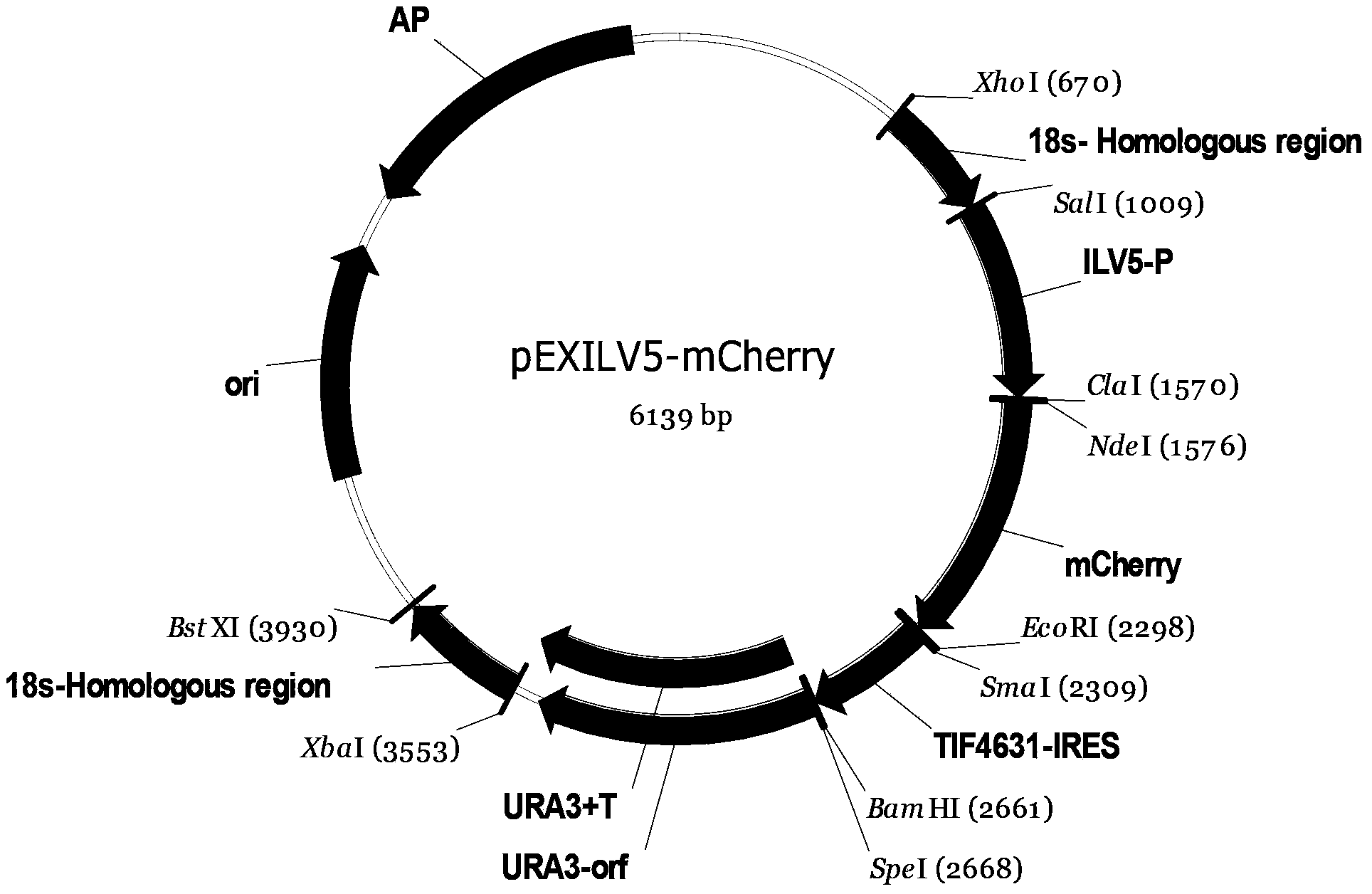
[0033] The preparation method of the plasmid pSK-mCherry is as follows: the DNA molecule shown in the 1568th to 2296th nucleotides from the 5' end of the sequence 1 of the artificially synthesized sequence listing (wherein the 1568th to 1573rd nucleotides are recognized by ClaI digestion) The sequence "ATCGAT", the 1574th to 1579th nucleotides are the NdeI enzyme digestion recognition sequence "CATATG", the 1580th to 2296th nucleotides are the coding gene of mCherry protein, and the 2297th to 2302th nucleotides are EcoRI digestion The recognition sequence "GAATTC") was inserted between the ClaI and EcoRI restriction sites of the pBlueScriptIISK(-) vector to obtain the plasmid pSK-mCherry.
[0034] pBlueScriptIISK(-) vector: purchased from Agilent Technologies Genomics, Catalog No. 212206.
[0035]Saccharomyces cerevisiae w303 strain (strain attributes: MATa; ura3-52; trp1Δ2; leu2-3,112; hi s3-11; ade2-1; can1-100): purchased from EUROSCARF, number 20000A.
Embodiment 1
[0036] Embodiment 1, the preparation of Saccharomyces cerevisiae expression vector
[0037] 1. Construction of recombinant plasmid pEXILV5-mCherry
[0038] 1. Extract the genomic DNA of Saccharomyces cerevisiae (the Latin name is Saccharomyces cerevisiae; China Center for General Microorganism Culture Collection and Management Center, the strain number is 2.1882).
[0039] 2. Using the genomic DNA extracted in step 1 as a template, use a primer pair (target sequence 554bp) consisting of ILV5P-S (underlined SalI recognition sequence) and ILV5P-AS (underlined ClaI recognition sequence) to perform PCR amplification to obtain PCR amplification product with ILV5 gene promoter of Saccharomyces cerevisiae.
[0040] ILV5P-S: 5'-ACGC GTC GAC CCCTATCTGTTCTTCCGCTCTACC-3';
[0041] ILV5P-AS: 5'-CC ATCGAT GTTTTATTTTTACTTATATTGCTGGTAGG-3'.
[0042] 3. Double-digest the PCR amplified product in step 2 with restriction enzymes SalI and ClaI, and recover the digested product.
[0043] ...
Embodiment 2
[0087] Example 2, Detection of the expression level of the Saccharomyces cerevisiae expression vector of the present invention
[0088] 1. Construction of Saccharomyces cerevisiae recombinant bacteria expressing mCherry
[0089] 1. Digest the recombinant plasmid pEXILV5-mCherry with restriction endonucleases XhoI and BstXI, and recover the digested product (with the upstream homology arm of 18SrDNA, the ILV5 gene promoter, the coding gene of mCherry protein, IRES sequence, URA3 gene, URA3 gene transcription termination sequence and downstream homology arm of 18S rDNA). The digested product was passed the lithium acetate method (Ito, H., Fukuda, Y., Murata, K., and Kimura, A.1983. Transformation of intact yeast cells treated with alkaline cations. J. Bacteriol. 53: 163-168 .) Transform Saccharomyces cerevisiae w303 to obtain a recombinant strain, which is named recombinant strain I-rfp.
[0090] 2. Digest the recombinant plasmid pEXTDH3-mCherry with restriction endonucleases ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com