Method for detecting 5-methylcystein in DNA
A technology of methylcytosine and cytosine, applied in the field of nucleic acid analysis, can solve problems such as ineffective reflection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1: Sodium bisulfite treatment of DNA
[0022] Dissolve less than 2 μg of DNA in 50 μl of ultrapure water, add 5.5 μl of freshly prepared NaOH (initial concentration: 3 M), and incubate in a 42° C. water bath for 30 minutes. Prepare 10 mM hydroquinone (hydroquinone) during the water bath. After the 30-minute water bath, add 30 μl to the DNA mixed solution, and the solution turns yellow. Then add 520 μl 3.6M NaHSO 3 , Gently invert the solution to mix well. Finally, add 200 μl of paraffin oil and react in a 50° C. water bath for 16 hours. After the reaction was completed, the mixture was desalted with a pore membrane, and precipitated with ice ethanol at -80°C for 2 hours, dried and dissolved in a certain amount of ultrapure water for UV quantification.
Embodiment 2
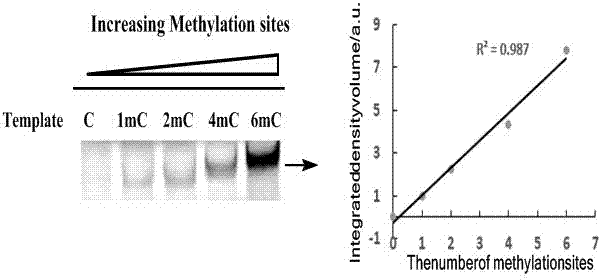
[0023] Example 2: Quantitative detection of 5-methylcytosine content by neutral polyacrylamide gel electrophoresis.
[0024] The upstream primer is GGGTTTTATTTATTTTAATTAATATTATATT (SEQ ID No 2), and the downstream primer is TCACCACTTCTCCCCTCAAT (SEQ ID No 3).
[0025] The asymmetric PCR reaction solution contains 1×PCR reaction buffer, 1μl dATP, dTTP and dCTP (initial concentration is 2mM), 0.5μl dGTP (initial concentration is 1mM), 1.5μl Fluorescein-dGTP (initial concentration is 1mM ), and finally ensure that the final concentration of these four bases is the same, which is 200 μM; 2.5U HotstartTaq polymerase, 0.5 μl upstream primer (initial concentration is 1 μM), 5 μl downstream primer (initial concentration is 10 μM) and sodium bisulfite treatment The final DNA template (20ng); the final reaction volume was 20μl. The reaction conditions for asymmetric PCR are: denaturation at 95°C for 15 minutes, followed by 35 amplification cycles (denaturation at 94°C for 30 seconds, a...
Embodiment 3
[0028] Example 3: Detection of the methylation level of the E-cadherin tumor suppressor gene promoter in the genomes of MDA-MB-231 and 97L cells by means of fluorescence.
[0029] Upstream primer TAGTAATTTTAGGTTAGAGGGTTAT (SEQ ID No 4), downstream primer AAACTCACAAATACTTTACAATTCC (SEQ ID No 5).
[0030] Both MDA-MB-231 and 97L cells were cultured in modified-1640 medium containing 10% fetal bovine serum, 1% streptomycin-penicillin, 5% CO 2 cultured in an incubator. Subsequently, the DNA of the two cell lines was extracted with a genomic DNA extraction kit, followed by sodium bisulfite treatment and asymmetric PCR. The conditions of asymmetric PCR are basically the same as in Example 2. 1×PCR reaction buffer, 1μl dATP, dTTP and dCTP (initial concentration is 2mM), 0.5μl dGTP (initial concentration is 1mM), 1.5μl Fluorescein-dGTP (initial concentration is 1mM), 2.5U Hotstart Taq polymerization Enzyme, 0.5 μl upstream primer (initial concentration is 1 μM), 5 μl downstream pri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com