Method for detecting drug-resistant mutation sites of campylobacter jejuni carbostyril antibiotics
A technology of campylobacter jejuni and mutant type, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve the problems of high cost, high culture conditions, and time-consuming, and achieve good specificity and sensitivity, The effect of accurate medication guidance and simplification of complex procedures
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
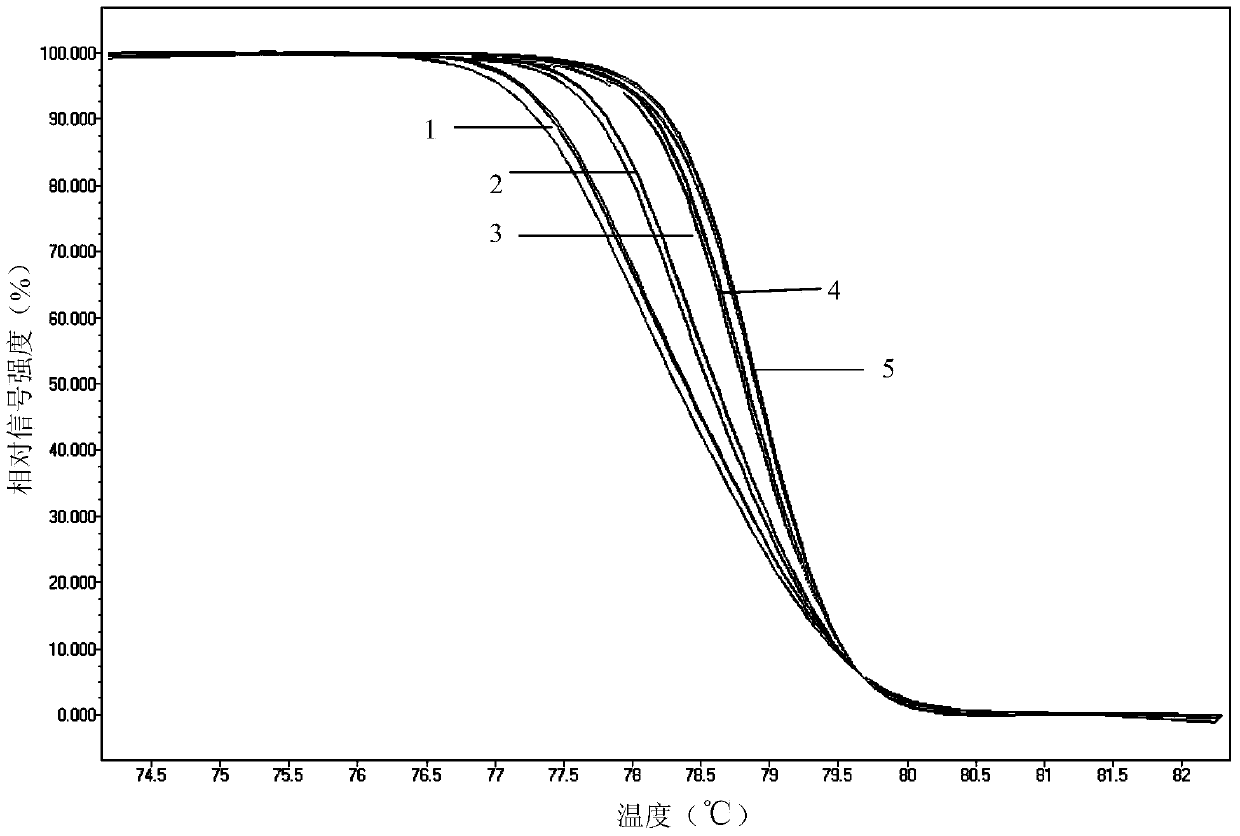
[0027] Example 1. Using high-resolution melting curve analysis to assist in the detection or detection of whether the 257th position of the gyrA gene coding sequence of Campylobacter jejuni is C or T
[0028] In this example, Roche LightCycler480 real-time fluorescent quantitative PCR system (the system includes Roche 480 fluorescent quantitative PCR instrument and Roche’s Gene Scanning module) was used for high-resolution melting curve analysis to establish auxiliary detection or detection of Campylobacter jejuni gyrA The method of whether the 257th position of the gene coding sequence is C or T. details as follows:
[0029] 1. Design of PCR amplification primers in high resolution melting curve analysis
[0030] jejuni NCTC11168 strain, Campylobacter jejuni ZP11, Campylobacter jejuni standard quality control strain ATCC33560, quinolone-resistant Campylobacter jejuni HT8, Campylobacter jejuni LY10, Campylobacter jejuni LY17, Campylobacter jejuni SX4, Campylobacter jejuni Co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com