Enol dehydrogenase, encoding gene, carrier, engineering bacteria and application of enol dehydrogenase
A technology of enol dehydrogenase and encoding gene, which is applied in the field of enol dehydrogenase and its encoding gene, can solve the problems of reducing the yield of target reaction products, side reactions, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Use the nucleic acid extraction kit to extract the genomic DNA of Yorkella sp.WZY002 (Yokenella sp. AGCTATGCCGCAAAAGAG-3'), primer 2 (5'-AAAGTCGGCTTGCAGTACC-3') for PCR amplification.
[0065] The amount of each component in the PCR reaction system (total volume 50 μL):
[0066] 10×TransStart rTaq Buffer (with Mg 2+ ) 5 μL, 10 mM dNTP mixture (2.5 mM each of dATP, dCTP, dGTP and dTTP) 12.5 μL, 1 μL each of primer 1 (1-1 or 1-2) and primer 2 at a concentration of 50 pM, 1 μL of genomic DNA, 1 μL of TransTaq DNA polymerase , add water to make up to 50 μL.
[0067] The PCR reaction conditions were as follows: pre-denaturation at 94°C for 4min, followed by a temperature cycle at 94°C for 30s; 55°C for 30s; 72°C for 1min; a total of 30 cycles, and a final extension at 72°C for 10min with a termination temperature of 4°C.
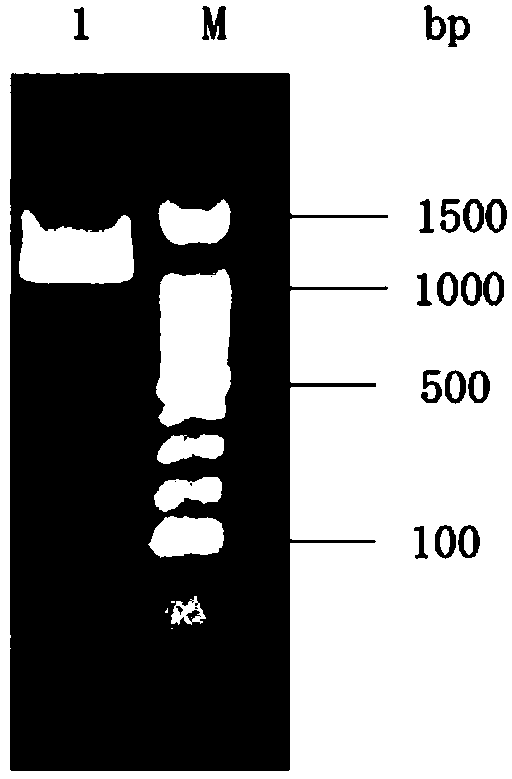
[0068] Take 6 μL of the PCR reaction solution and use 0.8% agarose gel electrophoresis to detect it. The electrophoresis result shows that it is a sing...
Embodiment 2
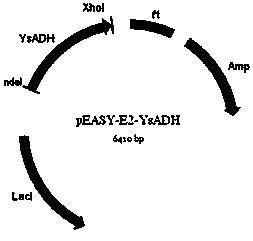
[0070]The recombinant plasmid obtained according to Example 1 was chemically transformed into Escherichia coli Trans B (DE3) (the competent cells were purchased from Beijing Quanshijin Biotechnology Co., Ltd.), and the recombinant plasmid pEASY-E2-YsADH containing intracellular expression was obtained Recombinant Escherichia coli Trans B(DE3) / pEASY-E2-YsADH. The recombinant Escherichia coli was cultured with LB liquid medium containing ampicillin (50 μg / ml) and kanamycin (50 μg / ml) at 37°C for 16 hours, and then inoculated with 1% inoculum (v / v) into fresh containing Ampicillin (50 μg / ml) and kanamycin (50 μg / ml) in LB liquid medium, cultured at 37°C until the cell concentration OD600 is about 0.6, and then added to the LB liquid medium with a final concentration of 0.2mM IPTG, induced at 22°C for 6-8 hours, then centrifuged at 10,000 rpm for 10 minutes at 4°C, and collected bacterial cells containing recombinant enol dehydrogenase. The obtained recombinant genetically engine...
Embodiment 3
[0075] The recombinant Escherichia coli Trans B (DE3) / pEASY-E2-YsADH wet cell was obtained in Example 2, and the cell was resuspended with 50mM Tris-HCl buffer solution of pH 8.0, and then ultrasonicated (sonication 2s, interval 6s , the effective ultrasonic time is 5min), the broken suspension is centrifuged at 10000rpm at 4°C for 10min, and the cell debris is discarded as much as possible to obtain the protein crude enzyme solution. According to the instructions of Ni-NTA metal chelate affinity chromatography, take the protein crude enzyme solution and load it on the pre-equilibrated Ni 2+ In the column, use 10mM imidazole, 40mM imidazole, 100mM imidazole, 250mM imidazole to elute the impurity protein and target protein in sequence. After the recombinant enol dehydrogenase was eluted with 100 mM imidazole, the obtained enzyme solution was desalted and concentrated by ultrafiltration membrane, and then stored at -20°C.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com