Novel proline specific endoprotease gene and application thereof
An endoprotease, proline technology, applied in the application, genetic engineering, plant gene improvement and other directions, can solve the problems of the proline specific endoprotease coding sequence and preparation method, etc., to improve beer abiotic Effects of stability, increased yield, and large application potential
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 Acquisition of A.oryzae proline-specific endoprotease gene
[0047] Based on Aeromonas (Genbank: AF065429), Pseudomonas capsular (Genbank: AB010298), Myxococcus flavum (Genbank: ABF89794) and Aspergillus fumigatus (Genbank: XM744168) and Aspergillus niger (Genbank: AX458699) sources The proline-specific endoprotease protein sequence design degenerate primers are as follows:
[0048] g1:5-RASMTWSRYATYASGGRA-3
[0049] g2:5-SVBYCBCYMBRGRKMANRNTB-3
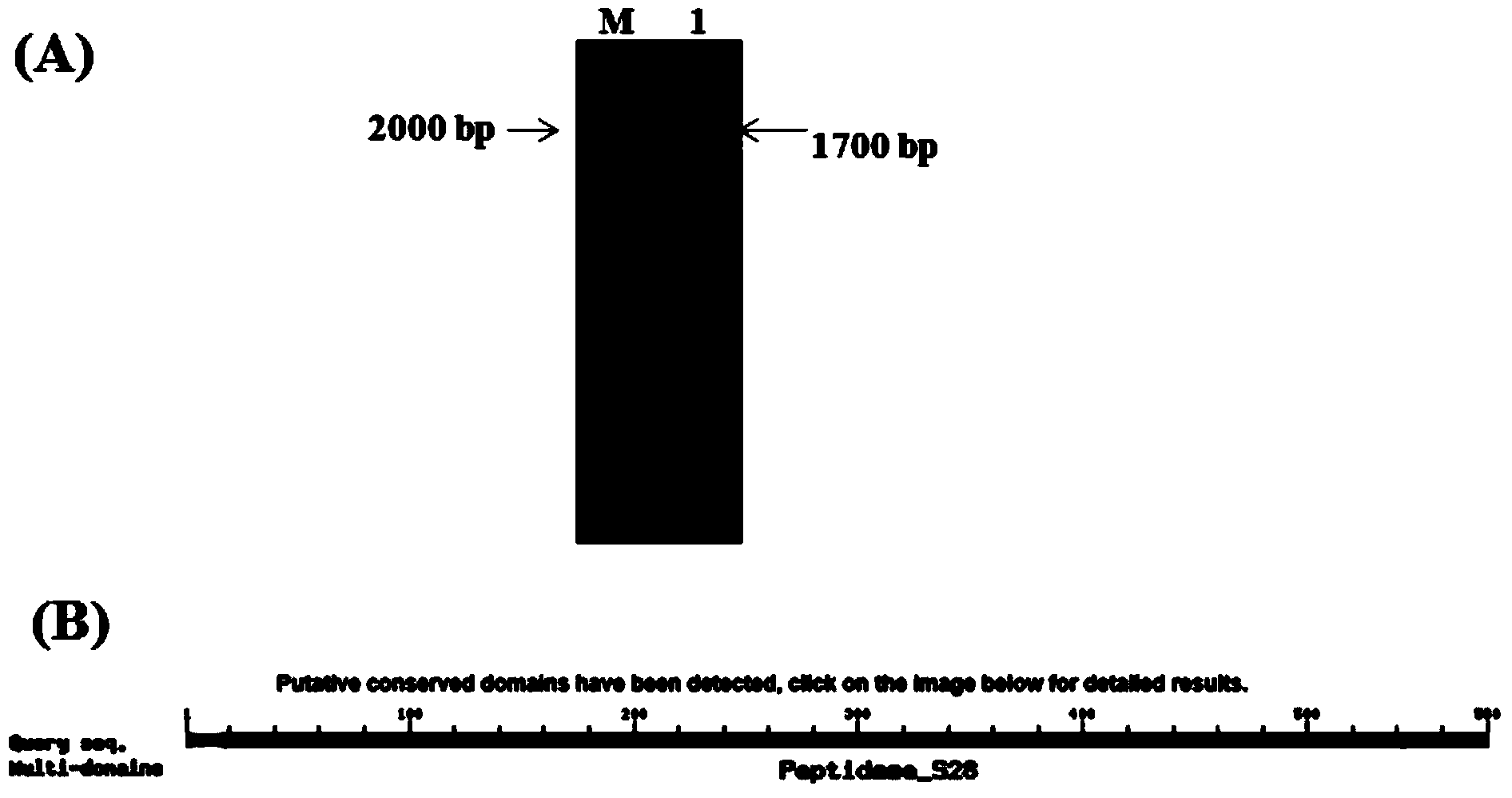
[0050] Using Aspergillus oryzae cDNA as a template and g1 and g2 as primers for PCR, a 1000bp product fragment was obtained. After the product was sequenced and compared with NCBI, it was found that it was all related to the A. oryzae protease gene (Sequence ID: XM_001825944.2 ) with high conserved sequence homology.
[0051] Furthermore, we redesigned primers and performed PCR based on the sequence of the A. oryzae protease gene (Sequence ID: XM_001825944.2) published in the GenBank database. The steps are as fol...
Embodiment 2
[0072] Example 2 Obtaining the Predicted Crystal Structure of A.oryzae Proline-Specific Endoprotease and Its Amino Acid Similarity with Other Aspergillus Proline-Specific Endoproteases Using "Homologous Modeling"
[0073] The total length of the Aspergillus oryzae proline-specific endoprotease gene S2 is 1743bp, and the predicted open reading frame of the new protein is located at nucleotides 64-1743, encoding 558 amino acid residues and a molecular weight of 65kDa.
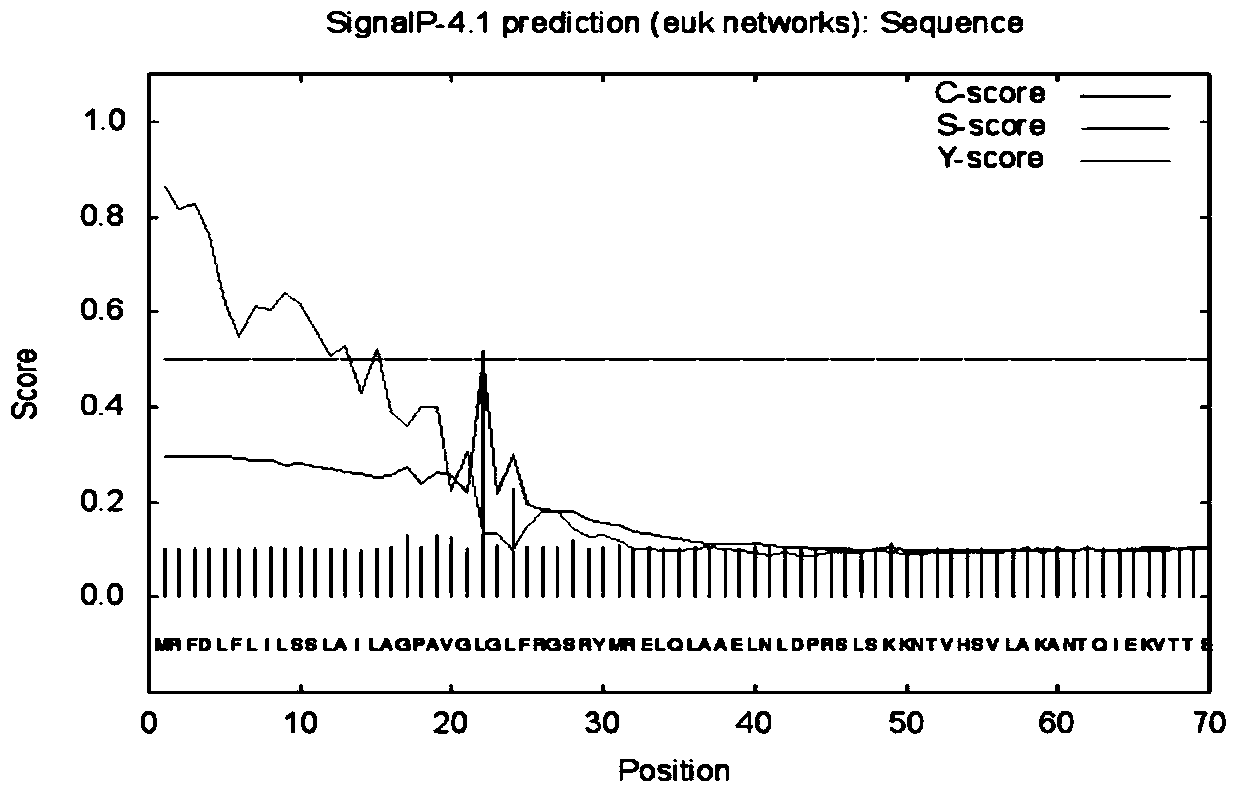
[0074] According to SignalP prediction, the possibility of the N-terminus of the protein being a signal peptide is 86.4%, and the cleavage site of the signal peptide is located between amino acids 21 and 22 (see figure 2 ).
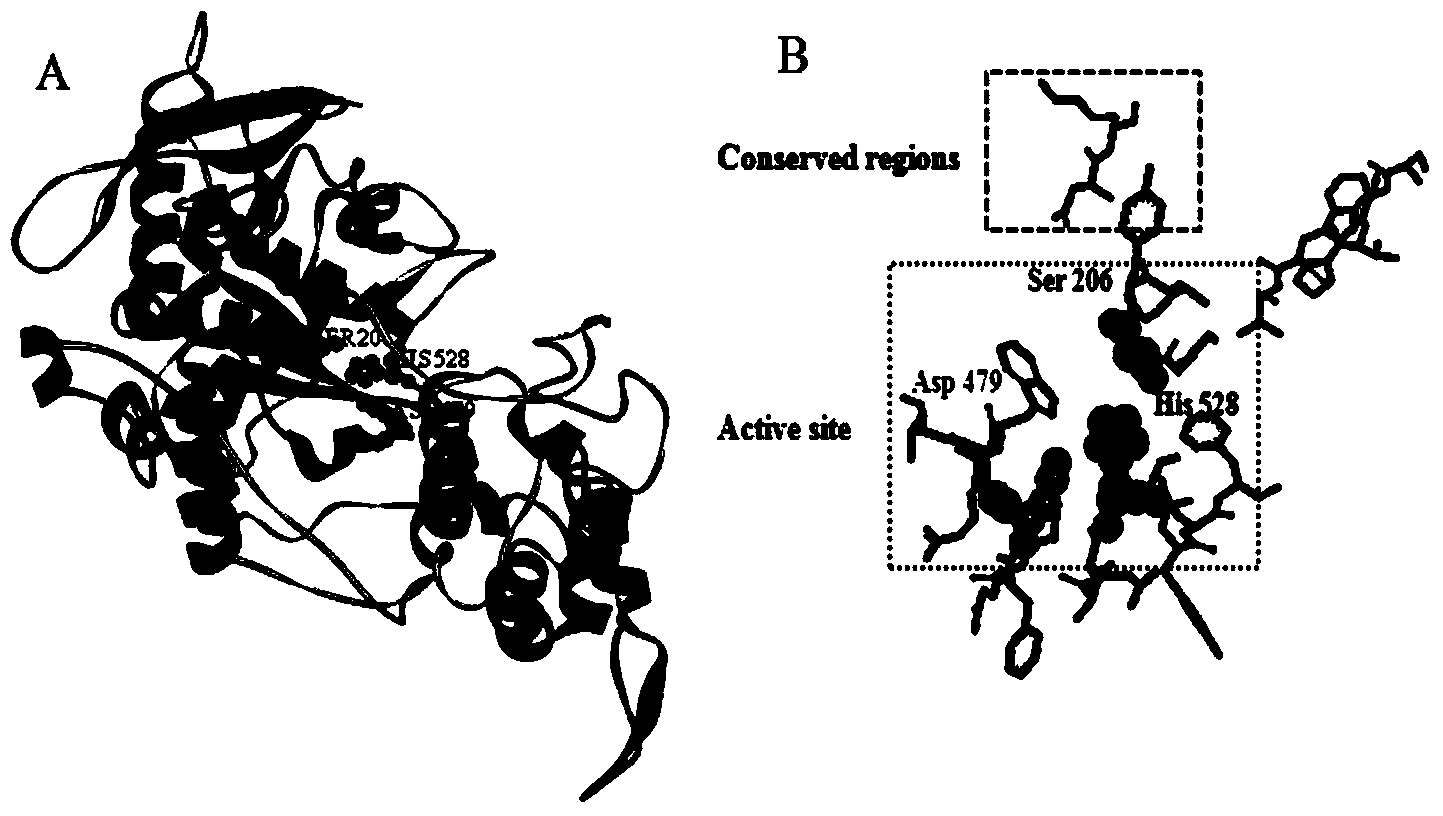
[0075]Submit the amino acid sequence of A. oryzae proline-specific endoprotease to the SWISS-MODEL protein online modeling server (http: / / swissmodel.expasy.org / ) for homology modeling, and then use Discovery studio software to analyze Aspergillus oryzae Proline-specific endoprotease protei...
Embodiment 3
[0077] Example 3 Construction of Aspergillus oryzae proline-specific endoprotease eukaryotic expression vector, recombinant expression and its protein expression
[0078] 1. Construction of eukaryotic expression vector
[0079] 1) Primer design: design primers starting from the mature peptide sequence after the signal peptide
[0080] g5:5′-CGG TACGTA TTGGGGT TGTTTAGAGG-3';
[0081] g6:5'-CC GCGGCCGC CTACATCACCGCCCCCTTTG-3';
[0082] 2) PCR reaction, using the cloning vector pMD-19T-S2 as a template, annealing at 62°C, 35 cycles.
[0083] 3) SnaBI and NotI double digestion PCR product of S2 and plasmid pPIC-9
[0084] Element
Usage amount
Purification of PCR products / plasmids
30μl
10*quitcut buffer
5μl
QuitCut SnaBI
1μl
Quit Cut Not I
1μl
wxya 2 o
13μl
total capacity
50μl
[0085] Digest at 37℃ for 2hr
[0086] 4) Ligate, transform, and identify by double enzyme digestion accordi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com