Graphene nanopore-microcavity-solid-state nanopore structure based DNA sequencing device and method
A graphene nanopore and DNA sequencing technology, applied in the field of DNA molecular sequencing, can solve the problems of sensitivity to environmental factors, low identification rate of base detection, short life, etc., and achieve the effect of improving low signal-to-noise ratio and improving sequencing accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0021] The present invention will be further described below in conjunction with the accompanying drawings and specific embodiments.
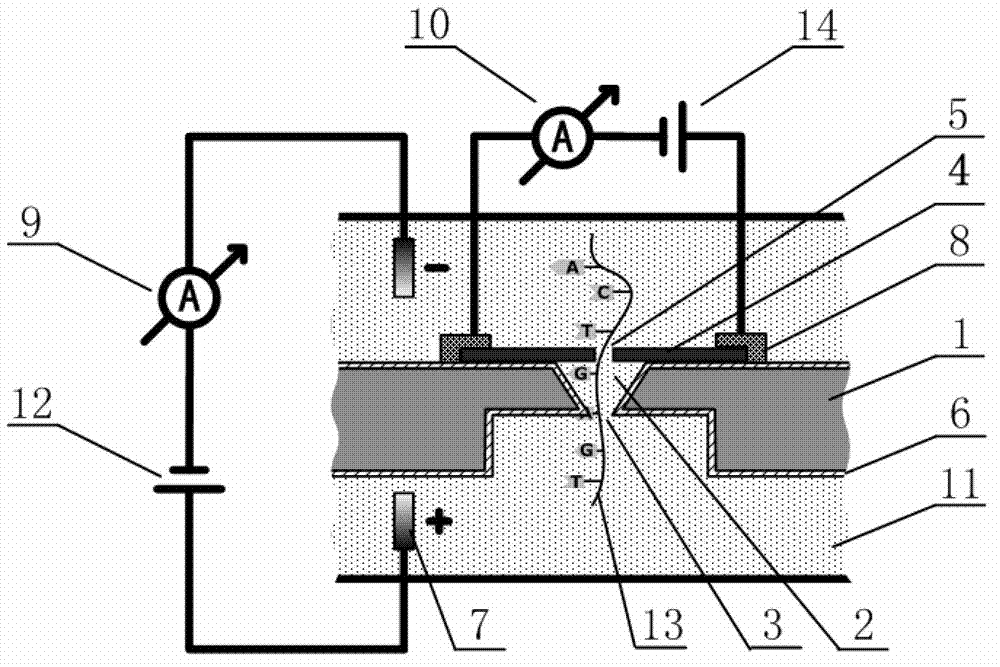
[0022] As shown in the attached figure, the DNA sequencing device based on the graphene nanopore-microcavity-solid-state nanopore structure, the DNA sequencing device is a sequencing device assembled with the graphene nanopore-microcavity-solid-state nanopore structure as the core, specifically Including a silicon-based substrate 1 placed in an electrolyte 11, an inverted pyramid-shaped microcavity 2 is etched on the upper half of the silicon-based substrate 1, and an inverted pyramid-shaped microcavity 2 is etched with a diameter larger than the bottom diameter of the inverted pyramid-shaped microcavity 2 in the lower half columnar holes, the top of the inverted pyramid-shaped microcavity 2 is a solid nanopore 3, the inverted pyramid-shaped microcavity 2 is used to control the speed of DNA molecular chains passing through the solid nanopore 3, ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com