Sequential separation and mass spectrum identification method of multi-site phosphorylation peptide
An identification method and a phosphorylation technology, which are applied in the direction of material analysis, material analysis, and measurement devices by electromagnetic means, can solve the difficulties in the identification of single phosphorylated peptides and the impossibility of single phosphorylation modification and multi-site phosphorylation modification. Distinguished, easy to break and other problems, to achieve the effect of simple and easy to control the operation process, low cost, and mild reaction conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
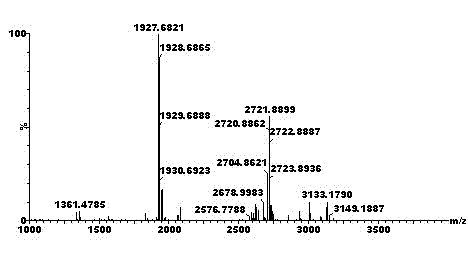
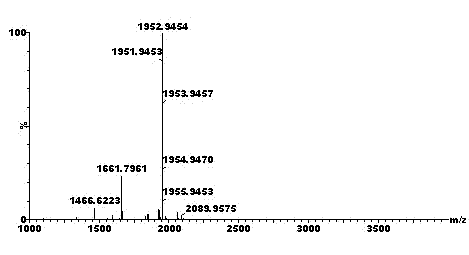
[0060] Sequential Separation of Multi-site Phosphorylated Modified Peptides and Sample Mass Spectrometry Analysis
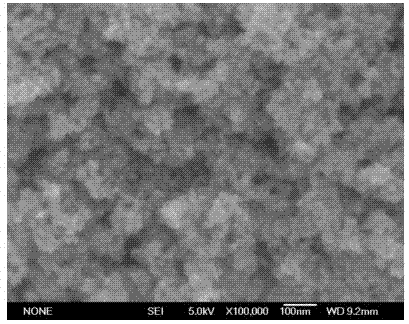
[0061] 1) Weigh 5~10 mg NiZnFe 2 o 4 Magnetic nano-ferrite material in a centrifuge tube;
[0062] 2), the NiZnFe 2 o 4 The magnetic nano-ferrite material is washed 3 times with a solution containing 50wt% acetonitrile and 0.1wt% TFA, separates the nanoparticles with a magnet, and discards the supernatant;
[0063] 3) Take the NiZnFe obtained in step 2) 2 o 4 The magnetic nano-ferrite particles were washed 3 times with 0.1wt% TFA aqueous solution, and the nanoparticles (NiZnFe 2 o 4 Magnetic beads), discard the supernatant;
[0064] 4) Before analyzing the samples, trypsinize the protein in a 0.1M ammonium bicarbonate solution at 37°C in a water bath for 12 hours;
[0065] 5) Adjust the pH of the enzymolysis solution obtained in step 4) to 1~2 with trifluoroacetic acid (TFA), add acetonitrile and TFA to make it contain 50wt% acetonitrile and 0.1wt% ...
Embodiment 2
[0072] The method of the present invention is used to identify phosphorylated proteins in zebrafish eggs
[0073] 1) Wash zebrafish egg cells with 0.675% saline, add to a glass tissue homogenizer, and mix with cell lysate (the lysate is a buffer solution composed of Tris-HCl and NaCl, and contains detergent 0.1% SDS and 0.5mM enzyme Inhibitor phenylmethylsulfonyl chloride PMSF) mixed.
[0074] 2) Determination of egg protein content by Bradford method;
[0075] 3) The protein disulfide bonds were reduced with dithiothreitol, and derivatized with iodoacetamide, and then trypsinized in a 37°C water bath with a mass ratio of protein:enzyme = 50:1;
[0076] 4) Preparation or purchase of magnetic nano-ferrite materials: NiZnFe 2 o 4 Magnetic beads and Fe 3 o 4 magnetic beads;
[0077] 5), the NiZnFe obtained in step 4) 2 o 4 Magnetic beads and Fe 3 o 4 The magnetic beads were washed 3 times with a washing solution containing 50wt% acetonitrile and 0.1wt% TFA, and then...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com