Method for rapidly detecting expression of ANS (Anthocyanidin Synthetase) genes from different sources in rape seed capsule and application thereof
A technology of gene expression and detection method, applied in the fields of biotechnology and molecular breeding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] (1) According to the published Arabidopsis ANS gene and rapeseed ANS gene sequence (Abrahams S, Lee E, Walker A R, Tanner G J, Larkin P J, Ashton AR. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J, 2003,35:624-636; Mingli Yan , Xianjun Liu, Chunyun Guan, Xinbo Chen, Zhongsong Liu. Cloning and expression analysis of anthocyanidin synthase gene homolog from Brassica juncea , Mol Breeding, 2011, 28:313–322), through bioinformatics analysis, design a pair of degenerate primers to amplify the full-length cDNA of ANS gene, black mustard (BB), cabbage (CC), Chinese cabbage (AA) , Ethiopian mustard (BBCC) and Brassica napus (AABB) seed coat cDNA were used as templates for PCR amplification. The amplified products were separated by agarose gel electrophoresis, recovered from the gel, connected to the pMD18-T vector, and then the vector was transferred to competent E. The...
Embodiment 2
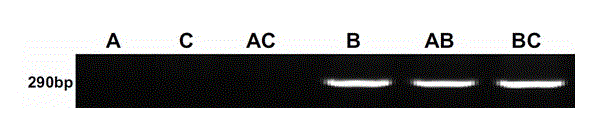
[0039] (1) Using cDNA as a template, PCR amplification was performed with specific primers (upstream primer 5'-TTAGCAAAGAGCGGAATCGAA-3', downstream primer 5'-CCACGGGCAAACCGAAGAA-3') from the ANS gene of the B chromosome group. The total volume of the PCR reaction system was 20 μL, including 2.0 μL of 10× PCR buffer, 0.3 μL of 10 mmol L?1 dNTP mix, 1 μL of 10 mmol L?1 forward primer, 1 μL of 10 mmol L?1 reverse primer, 1 U Taq DNA polymerase, 2.5 mmol L?1 MgCl2 2 μL, 1 μL reverse transcription product, sterile deionized water (make up to 20 μL). PCR amplification conditions were as follows: pre-denaturation at 94°C for 3 min; denaturation at 94°C for 25 s, annealing at 60°C for 30 s, extension at 72°C for 20 s, and 30 cycles; finally, extension at 72°C for 6 min.
[0040] (2) Amplified products were separated by 1.5% agarose gel electrophoresis and stained with ethidium bromide. Then take pictures and analyze through the gel analysis system, the results are as follows figu...
Embodiment 3
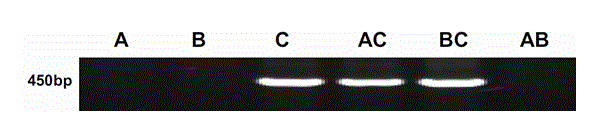
[0043] (1) Using cDNA as a template, PCR amplification was performed with specific primers from the ANS gene of the C chromosome group (upstream primer 5'-TAGCAATGGGAAGTTTAAGAGTA-3', downstream primer 5'-GTGGTTCATAGAGCATATCTCG-3'). The total volume of the PCR reaction system was 20 μL, including 2.0 μL of 10× PCR buffer, 0.3 μL of 10 mmol L?1 dNTP mix, 1 μL of 10 mmol L?1 forward primer, 1 μL of 10 mmol L?1 reverse primer, 1 U Taq DNA polymerase, 2.5 mmol L?1 MgCl2 2 μL, 1 μL reverse transcription product, sterile deionized water (make up to 20 μL). PCR amplification conditions were as follows: pre-denaturation at 94°C for 3 min; denaturation at 94°C for 25 s, annealing at 57°C for 30 s, extension at 72°C for 30 s, and 30 cycles; finally, extension at 72°C for 6 min.
[0044] (2) Amplified products were separated by 1.5% agarose gel electrophoresis and stained with ethidium bromide. Then take pictures and analyze through the gel analysis system, the results are as follows i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com