Micro ribonucleic acid (RNA), and use of precursor and antisense nucleic acid thereof in regulation of catalase activity
A technology of catalase and RNA precursor, which is applied in the professional field of biomedicine and can solve problems such as increased expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
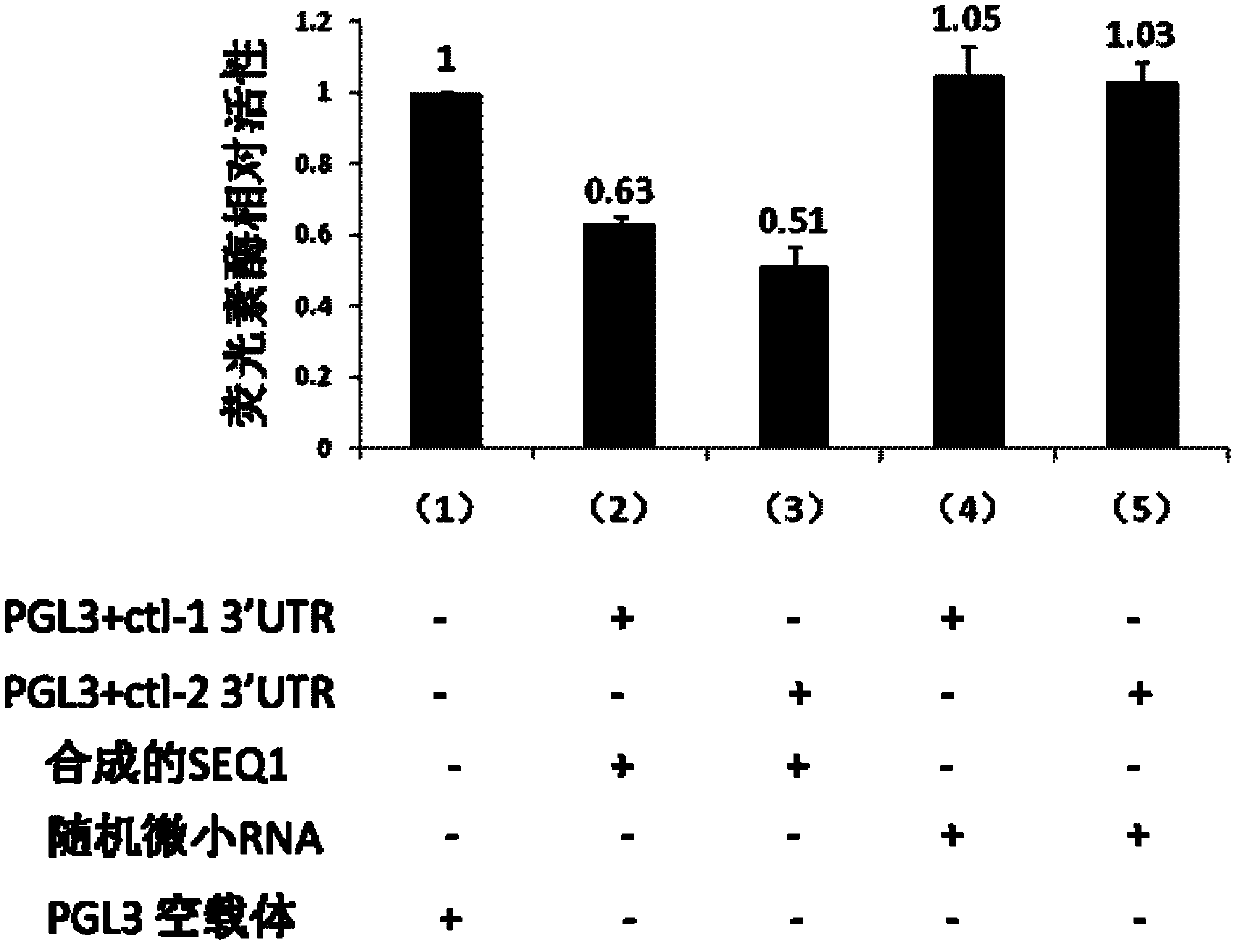
Examples
Embodiment 1
[0040] Cloning and analysis of embodiment 1miRNA
[0041] 1. Extraction and purification of RNA
[0042] Get 20 wild-type Caenorhabditis elegans, 20 SEQ1 deletion nematodes SEQ1 (- / -) respectively placed in 40ulM9 buffer (KH 2 PO 4 3g; Na 2 HPO 4 6g; NaCl5g; 1MMgSO 4 1ml; add water to make up to 1000ml. ) into a RNase-free 1.5ml centrifuge tube (purchased from Axygen), add 500ul of total RNA extraction reagent Trizol (purchased from Invitrogen), vortex for 30 seconds to mix, and ice-bath for 30 minutes to dissolve the parasites. This was followed by centrifugation at 12000 rpm for 10 minutes at 4°C. Transfer the supernatant to another RNase-free 1.5ml centrifuge tube, add 100ul chloroform, vortex for 30 seconds, and place at room temperature for 3 minutes. Centrifuge at 12000 rpm for 15 minutes at 4°C. Transfer the supernatant to another RNase-free centrifuge tube. Add an equal volume (usually about 300ul) of isopropanol and mix well. Let stand at room temperature fo...
Embodiment 2
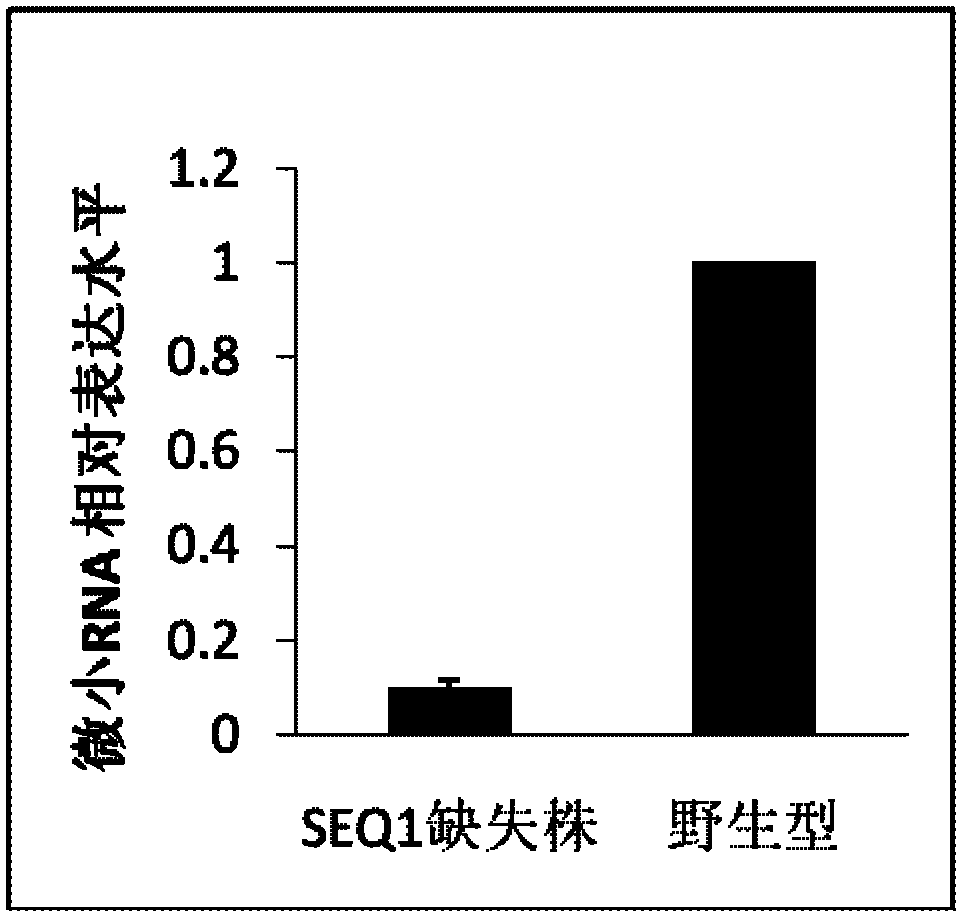
[0058] Embodiment 2 Real-time quantitative fluorescent PCR detects the expression of SEQ1 microRNA in nematode strains
[0059] Express
[0060] Wild-type Caenorhabditis elegans and SEQ1-deficient nematode strain SEQ1(- / -) were cultivated, and nematode RNA was extracted and reverse-transcribed according to the method in Example 1. After reverse transcription, the expression difference of SEQ1 between wild type and SEQ1(- / -) was detected by real-time quantitative fluorescent PCR (RT-PCR).
[0061] The SYBRPremixExTaq kit (purchased from TAKARA) was used according to the manufacturer's product instructions. Add 2ul template, 10ul2×SYBRPremixExTaq, 0.4ul forward primer (5'-GTGCAGGGTCCGAGGT-3') (SEQ.ID.No.4) 10uM, 0.4ul reverse primer (5'- AGGCAAGATGTTGGCA-3') 10uM, 0.4ul 50×ROXReference dye (included in the SYBRPremixExTaq kit), add sterilized distilled water to 20ul.
[0062] Use U6 as an internal reference, refer to the calculation method of QuantitativeReal-TimeRT-PCR of Th...
Embodiment 3
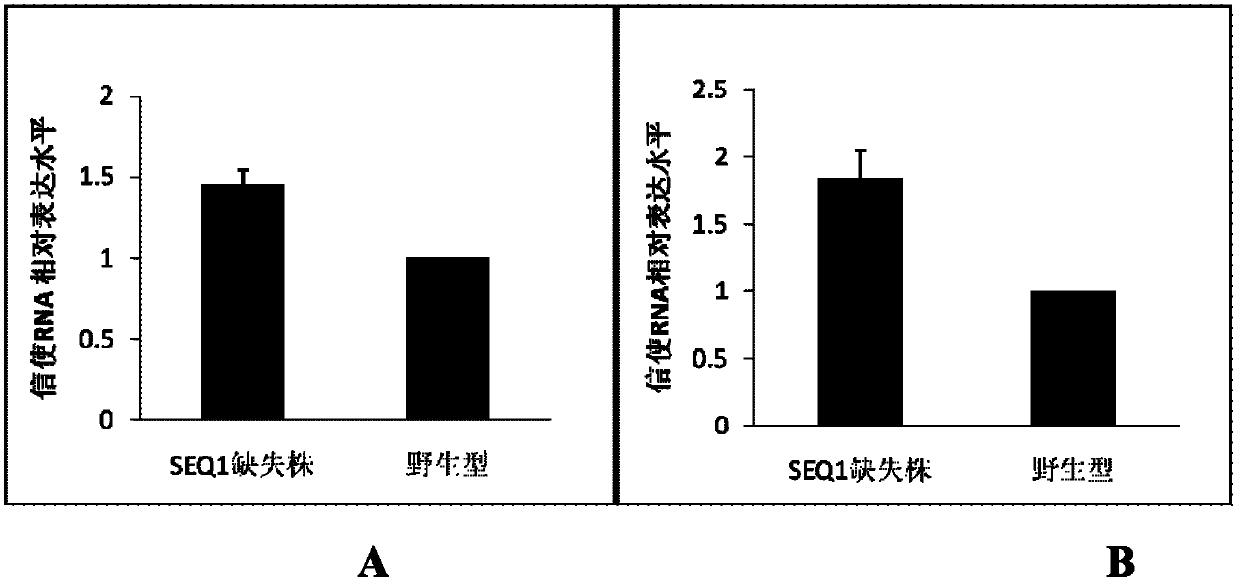
[0064] Example 3 Real-time quantitative fluorescent PCR detects the expression of catalase in each strain of nematode
[0065] Wild-type C. elegans and the SEQ1-deficient nematode strain SEQ1(- / -) were cultivated, and nematode RNA was extracted and reverse-transcribed according to the method in Example 1.
[0066] ctl-1 reverse transcription primer: 5'-GAGCTGATGGCGAAGAGC-3' (SEQ.ID.No.9)
[0067] ctl-2 reverse transcription primer: 5'-TGGATGAGTGCCTTGACA-3' (SEQ.ID.No.10)
[0068] After reverse transcription, real-time quantitative fluorescent PCR was used to detect the expression difference of ctl-1 and ctl-2 between wild type and SEQ1(- / -). The experimental conditions are the same as in Example 2.
[0069] Primers used in real-time quantitative fluorescent PCR reaction
[0070] ctl-1RT upstream primer: 5'-CGCATCTCCCTGGCTTTCAT-3' (SEQ.ID.No.11)
[0071] ctl-1RT downstream primer: 5'-CTCGCCAGACAAGATGCTCC-3' (SEQ.ID.No.12)
[0072] ctl-2RT upstream primer: 5'-ATCATGACATTCCC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com