Screening and verifying of human papilloma virus (HPV) 16 tripeptide vaccine and construction of tumor animal model for continually expressing HPV16 E5, E6 and E7
A HPV16E5, animal model technology, applied in anti-tumor drugs, viral antigen components, peptides, etc., to prevent tumor formation, inhibit invasion, and inhibit tumor formation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
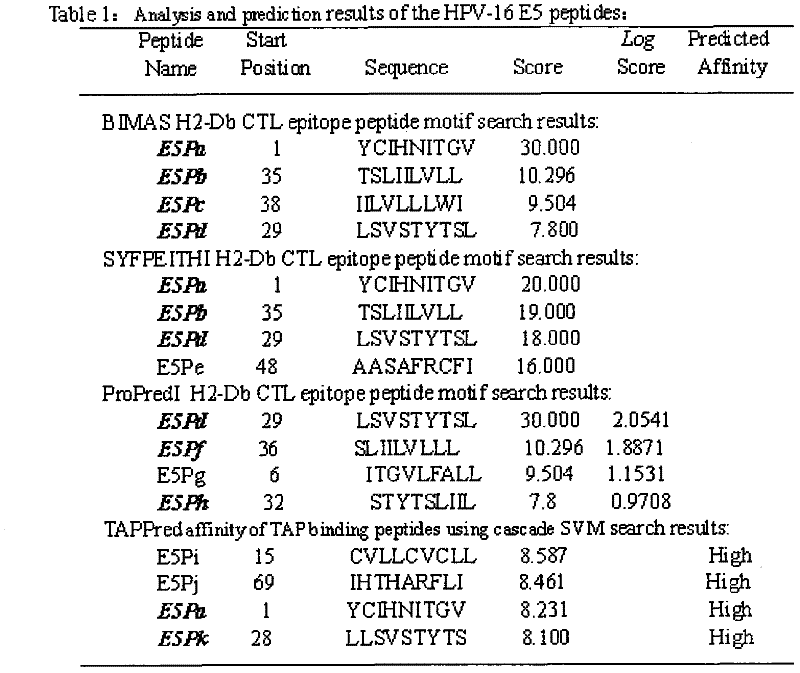
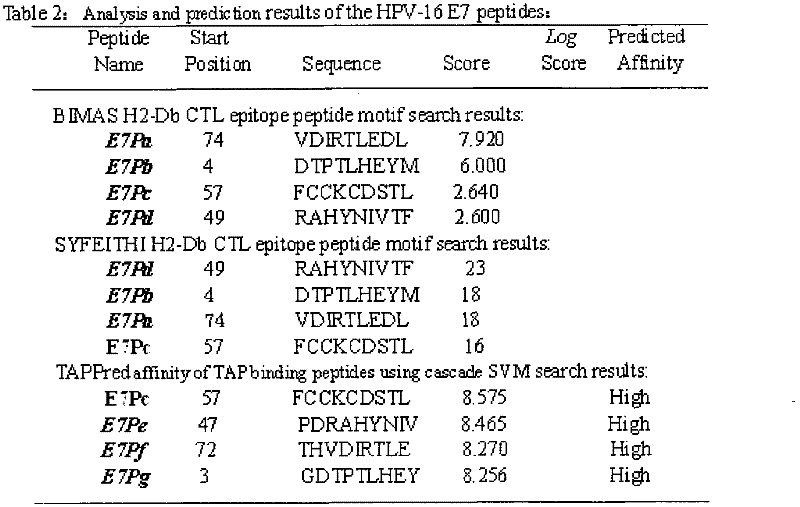
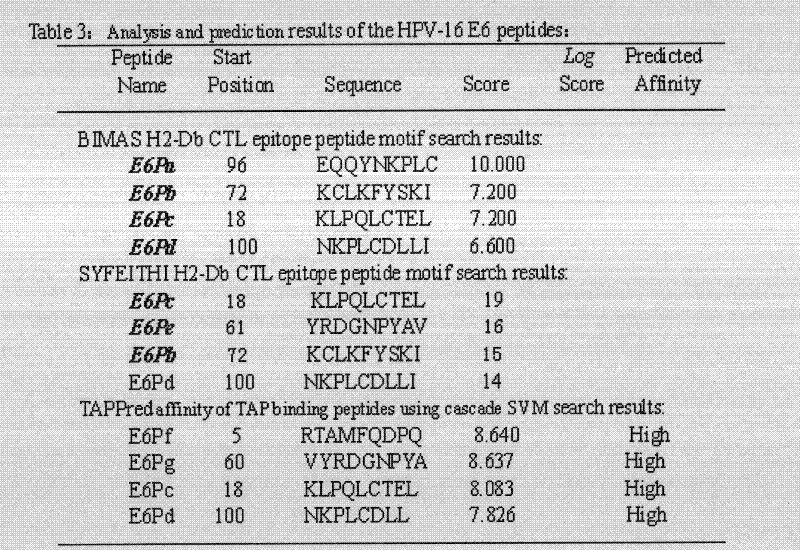
[0021] Example 1: Screening of HPV16 E5, E6, E7 polypeptide fragments
[0022] The full-length sequences of HPV16 E5, E6 and E7 were analyzed through the National Institute of Health Bioinformatics and Molecular Structure Analysis System, Antigen Epitope Prediction System and TAP Prediction System, and the corresponding fragments were obtained. We selected higher scores, strong affinity, and immune sources Target fragments with better sex and antigenicity were used as candidate fragments for further laboratory screening. For details, see (http: / / bimas.dcrt.nih.gov / cgi-bin / molbio / ken_parker_comboform) [1] (http: / / www.syfpeithi.de / ) [2] (http: / / www.imtech .res.in / cgibin / tappred / ) [3] See the results Figure 1-3
[0023] 1. Parker, K.C., M.A. Bednarek, and J.E. Coligan. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 1994; 152: 163-175.
[0024] 2. Rammensee, H.G., J.Bachmann, N.P.N.Emmerich, et a...
example 2
[0026] Example 2: Construction of tumor animal models that continuously express HPV16 E5, E6, and E7
[0027] 1) Construction of AAV-E5 destination vector
[0028] pEGFP-C1-HPV16 E5 and pAAV-MCS were digested with restriction endonuclease EcoR Ⅰ and BAMH Ⅰ respectively to construct AAV-E5 recombinant plasmid.
[0029] ① Enzyme digestion system
[0030]
[0031] Mix well, 37 ℃, 2-4h or overnight, electrophoresis analysis and identification.
[0032] ② Gel purification: (U-gene kit, see Part 2.2.23 for specific operation)
[0033] ③Connection:
[0034]
[0035] Mix well, overnight at 4°C, and identify by electrophoresis analysis.
[0036] ④ Transformation screening: transform the competent bacteria DH 5a with the ligation product. Coat the plate (kanamycin / IPTG / X-gel), carry out the blue and white spot screening, and select the white spot.
[0037] ⑤ Enzyme digestion identification: After extracting the plasmid DNA (Plasmid mini-extraction kit from Biotech, strictly ...
example 3
[0079] Example 3: Further screening and identification of HPV16 E5, E6, E7 polypeptide fragments by in vivo and in vitro tests.
[0080] (1) Tumor growth
[0081] First, the mice were randomly divided into 9 groups, and rTC-1 cells in the logarithmic growth phase were taken from each group to prepare single-cell suspensions (1×10 6 / 100μl), inoculate 200μl at the root of the left thigh of each mouse, and give the corresponding peptide preparation to the root of the left thigh of the mouse one week later, CpG and saline, a total of 9 groups, after that, give the corresponding peptide preparation once a week, or CpG, or saline. Once every 1-2 days, observe the formation of tumor masses. After a tumor grows, observe and measure the tumor size 2-3 times a week (vertically measure the longest diameter of the tumor a, b) until the tumor is formed or 3-4 weeks after treatment, calculated by the formula 1 / 2*a*b tumor volume.
[0082] (2) ELISA method to detect the expression of im...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com