Chlamydomonas reinhardtii lipid metabolism gene CrDGAT2-5, encoding protein thereof, and application thereof
A technology of Chlamydomonas reinhardtii, encoding protein, applied in the field of microorganisms, can solve the problem that there is no DGAT gene family yet
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
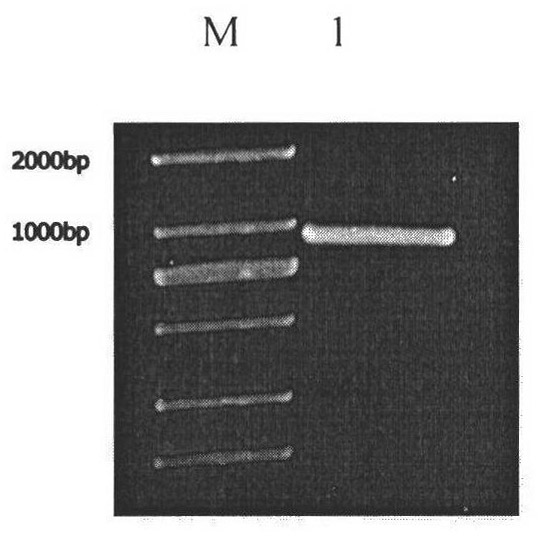
[0028] Example 1: Cloning of CrDGAT2-5
[0029] 1. Extraction of total RNA from Chlamydomonas reinhardtii
[0030] Chlamydomonas reinhardtii (Chlamydomonas reinhardtii) strain CC425, cultivated in a light incubator at 24°C, 100 μmol m -2 sec -1 Under white light and full light, after growing to the logarithmic growth phase, take 50 mL of the algae liquid, centrifuge at 10,000 r / min for 1 min to collect the algae, freeze them in liquid nitrogen, grind them into powder, and extract total RNA according to the method of Trizol.
[0031] 2. Primer design
[0032] According to the au.g4218_t1 gene sequence published on the JGI Chlamydomonas database as a template, the following specific primers were designed for PCR amplification:
[0033] CrDGAT2-5 full-length amplification primer Primer sequence (5'→3')
[0034] Forward primer ATGGCCTCTTACTTCCCCGGC
[0035] Reverse primer TCATTGCACGATGGCCAGCGG
[0036] 3. Synthesis of cDNA
[0037] According to Takar...
Embodiment 2
[0048] Example 2: Obtaining transformants overexpressing the CrDGAT2-5 gene
[0049] 1. Construction of plant binary expression vector
[0050] Design primers with NcoI and SpeI restriction sites (forward:
[0051] 5'-AGAACCATGGCAGGTGGAAAGTCAA ACGG-3'; Reverse:
[0052] 5'-CATAACTAGTCTACTCGATGGACAGCGGG-3'), the full-length CrDGAT2-5 gene was amplified by PCR. The CrDGAT2-5 product modified by NcoI and SpeI double digestion was ligated with the NcoI and SpeI double digestion vector large fragment of pCAMBIA1302. The ligation product was transformed into Escherichia coli DH5α, the positive clone was identified by PCR, and the plasmid was extracted for double enzyme digestion identification. The correct recombinant expression plasmid pCAMBIA1302-CrDGAT2-5 was extracted for transformation.
[0053] 2. Acquisition of overexpressed CrDGAT2-5 Chlamydomonas reinhardtii transformants (electrotransformation method)
[0054] Chlamydomonas reinhardtii (algae strain CC425, purchased f...
Embodiment 3
[0057] Example 3: Physiological phenotype observation of CrDGAT2-5 overexpressed algal strains
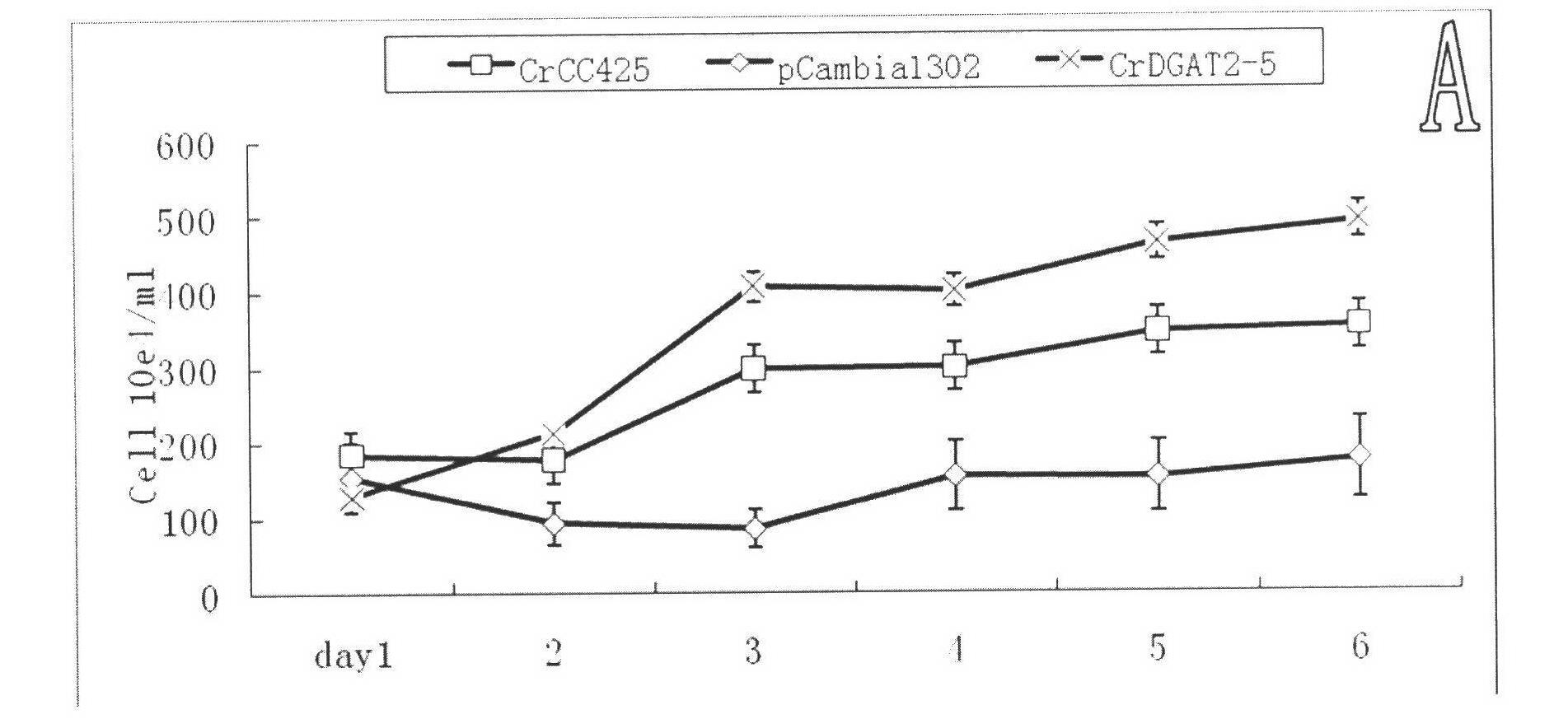
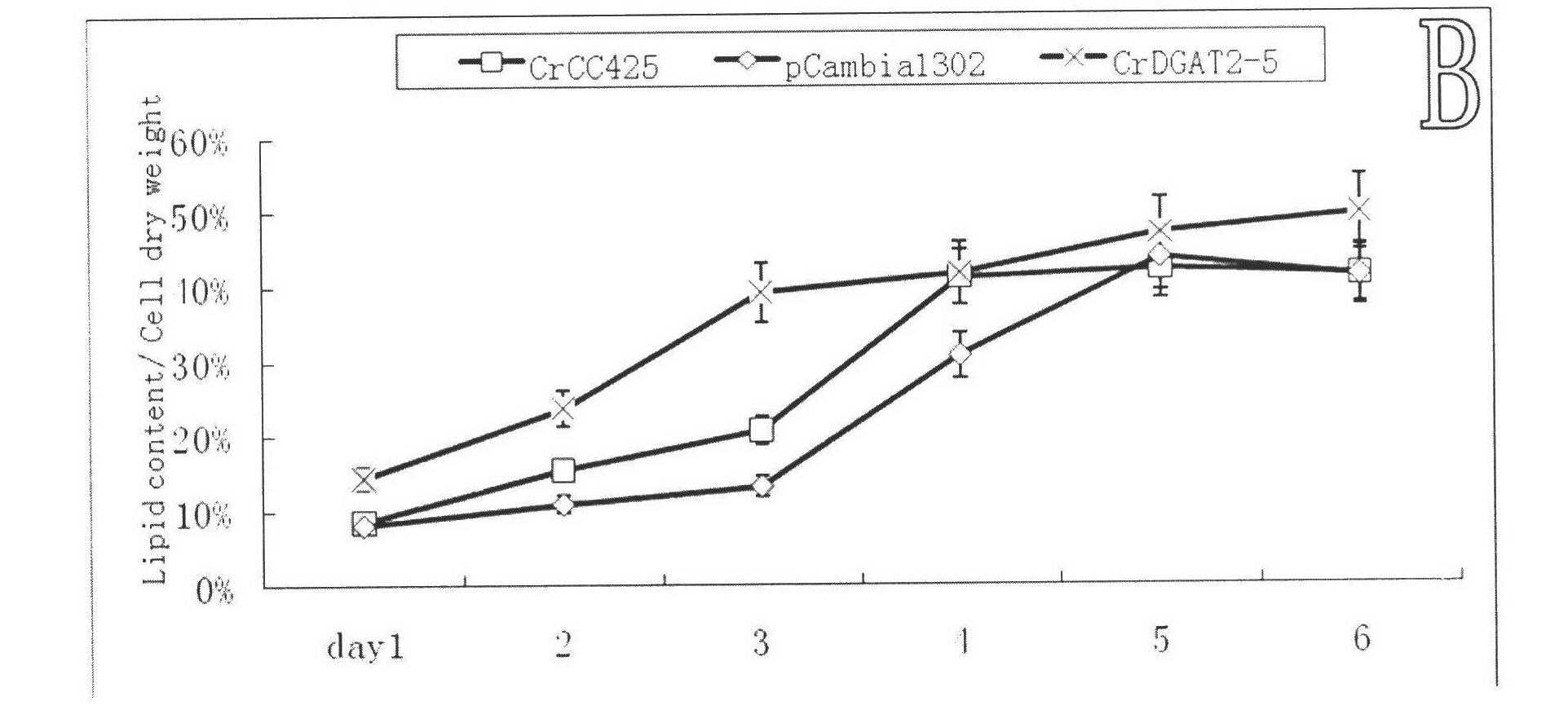
[0058] 100 algae strains were selected from the pCMBIA1302 empty vector group, CrDGAT2-5 overexpression group, and original algae strain CC425, and cultured in a light incubator at 24°C, 100 μmol m -2 sec -1 White light, under full light conditions, wait until the logarithmic growth phase, 2000rpm, centrifuge for 5min, resuspend in double distilled water, inoculate HSM-N liquid medium (2g / LNaAc 3H 2 O+1.44g / L K 2 HPO 4 +0.72g / L KH 2 PO 4 +0.5467g / L NaCl+20mg / LMgSO 4 ·7H 2 O+10mg / LCaCl 2 2H 2 O+1ml / L Trace-N), cultured with continuous shaking for 6 days. The following physiological values were measured every 24h.
[0059] 1. Biomass statistics
[0060] Biomass was calculated using a haemocytometer as described by Harris, 1989. The results showed that the algal strains overexpressing CrDGAT2-5 gene significantly increased the biomass of the non-transgenic strain CrCC425...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com