Open-source method for screening zinc finger proteins targeted combined with target sites of human DYRK1A gene
A zinc finger protein, targeted binding technology, applied in the field of genetic engineering, can solve the problems of limited zinc finger library capacity, inability to triple base screening, inability to achieve screening, etc., and achieve high efficiency, specificity and high affinity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] 1. Construction of 3 zinc finger protein libraries:
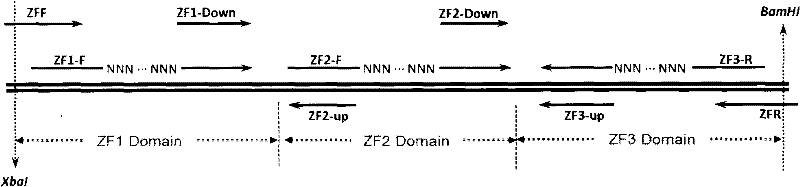
[0038] 1. ZF1 (zinc finger 1, zinc finger domain 1) random ZFP1 library construction
[0039] 1.1. Construction of random ZFP1 fragments of ZF1
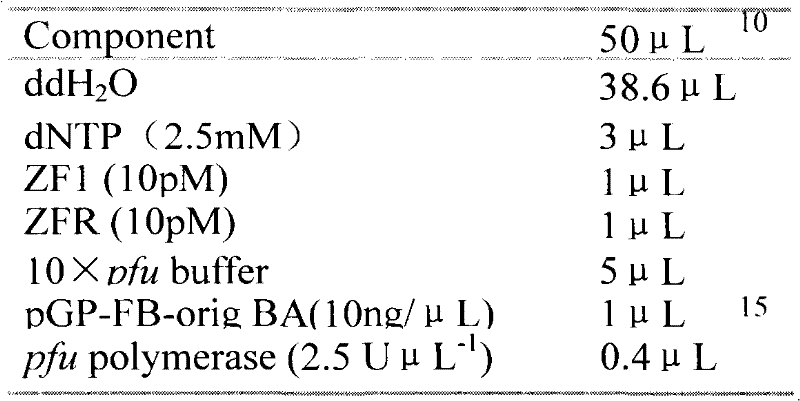
[0040] Using the zinc finger protein expression plasmid pGP-FB-orig BA as the substrate, ZF1-F and ZFR as primers, use pfu polymerase to amplify according to the PCR reaction system shown in Table 1. The cycle conditions are: 94°C for 5min, 94°C for 30s, 55°C for 30s, 72°C for 30s, a total of 28 cycles, and finally 72°C for 10min. Randomize the 21 key bases in ZF1, and then use this PCR reaction product as a substrate, ZFF and ZFR primers, use pfu polymerase according to the table The PCR reaction system shown in 2 was amplified, and the cycle conditions were: 94°C for 5 minutes, 94°C for 30s, 55°C for 30s, 72°C for 30s, a total of 20 cycles, and finally 72°C for 10 minutes to amplify the complete zinc finger protein expression fragment, namely ZF1 random ZFP1 fragment (R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com