Living cell detection method based on chemiluminescence principle
A chemiluminescence and detection method technology, applied in chemical instruments and methods, chemiluminescence/bioluminescence, luminescent materials, etc., can solve the problems of inability to measure ATP, fast detection speed, and high accuracy, so as to shorten detection time and reduce detection. Time, the effect of rapid detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
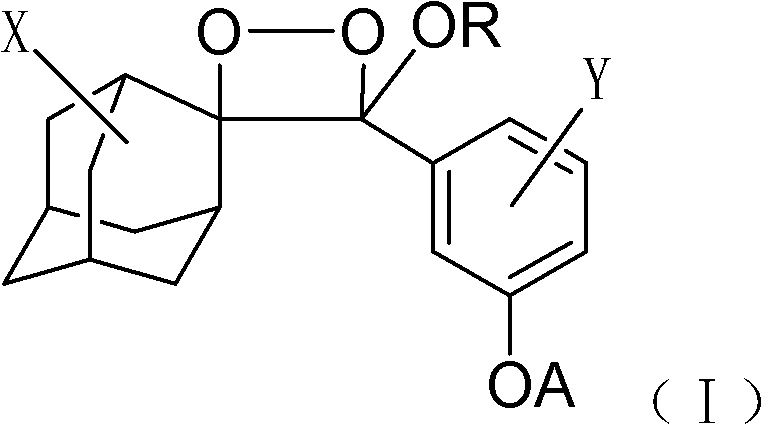
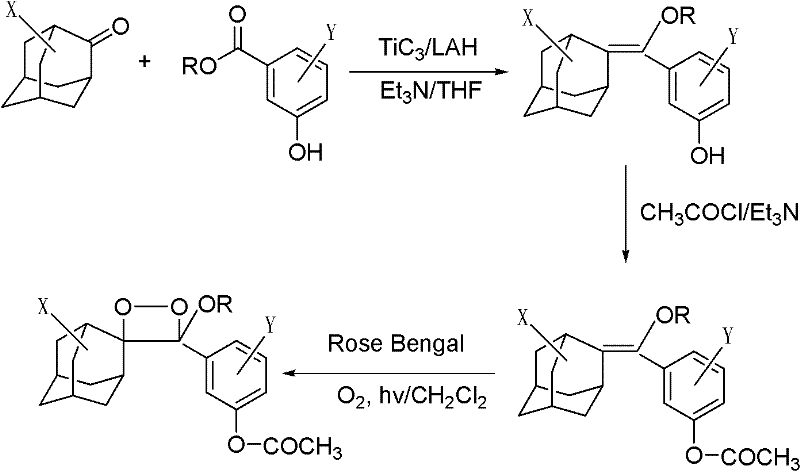
[0025] 1) Dissolve 1 mg of the phosphatase substrate (I-1) (R is octyl, X is the 5-position chlorine element, Y is the ortho-position chlorine element) with 1 ml of anhydrous DMSO to prepare a 1 mmol / l phosphatase substrate reserve solution, sealed and frozen at 2-8°C; add 10 μl of phosphatase substrate stock solution to 5 ml of Tris buffer to prepare 2 μmol / l, 0.2% DMSO phosphatase substrate solution.
[0026]
[0027] 2) Centrifuge the artificial Escherichia coli bacterial liquid of known concentration to collect the bacterial colony, remove the supernatant, add Tris buffer solution (pH8.0), and dilute 10 times into a series of bacterial liquid standards (the number of bacteria is adjusted to 10 4 、10 5 、10 6 、10 7 pieces / ml).
[0028] 3) Pipette 0.2ml of the Escherichia coli liquid with different concentrations above, transfer to a 96-well polystyrene microwell plate, add 0.2ml of the above phosphatase substrate solution, stir well and incubate at room temperature for...
Embodiment 2
[0031] 1) Dissolve 1 mg of lipase substrate (I-2) in 1 ml of anhydrous DMSO (R is a methyl group, X is a 5-position chlorine element, Y is an ortho-position chlorine element), and prepare a 1 mmol / l lipase substrate reserve Liquid, sealed and frozen at 2-8°C. Add 10 μl of lipase substrate stock solution to 5 ml of Tris buffer to prepare a 2 μmol / l, 0.2% DMSO lipase substrate solution.
[0032]
[0033] 2) Centrifuge the Pichia pastoris GS115 bacterial liquid of known concentration to collect the bacterial colony, remove the supernatant, add Tris buffer (pH8.5), and dilute 10 times into a series of bacterial liquid standards (adjust the number of yeast to 10 4 、10 5 、10 6 、10 7 pieces / ml).
[0034] 3) Take 0.2ml of the above-mentioned yeast liquids with different concentrations, transfer them to a 96-well polystyrene microwell plate, add 0.2ml of the above-mentioned lipase substrate solution, stir well and incubate at room temperature for 15 minutes to trigger chemilumin...
Embodiment 3
[0037]1) Dissolve 1 mg of lactase substrate (I-3) (R is ethyl, X is 5-position chlorine element, Y is ortho-position chlorine element) with 1 ml of anhydrous DMSO to prepare 1 mmol / l lactase substrate reserve Liquid, sealed and frozen at 2-8°C. Add 10 μl of lactase substrate stock solution to 5 ml of Tris buffer to prepare a 2 μmol / l, 0.2% DMSO lactase substrate solution.
[0038]
[0039] 2) Centrifuge the lactic acid bacteria liquid of known concentration to collect the flora, remove the supernatant, add Tris buffer (pH8.0), and prepare a certain concentration of the bacterial liquid sample (the number of lactic acid bacteria is adjusted to 10 5 and 10 6 pieces / ml).
[0040] 3) Take 0.2ml of the above-mentioned lactic acid bacteria solution of different concentrations, transfer it to a 96-well polystyrene microwell plate, add 0.2ml of the above-mentioned lactase substrate solution, stir well and incubate at room temperature for 15 minutes to trigger chemiluminescence, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com