System and method for high resolution of nucleic acids to detect sequence variations
A variation and sequence technology, applied in the field of detection of sequence variation related to drug resistance and/or sensitivity, can solve the problems of ignoring sequence variation and limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0189] Example 1 -Amplification and analysis of drug-sensitive regions of Mycobacterium tuberculosis
[0190] A. Whole-genome amplification of Mycobacterium tuberculosis genomic DNA
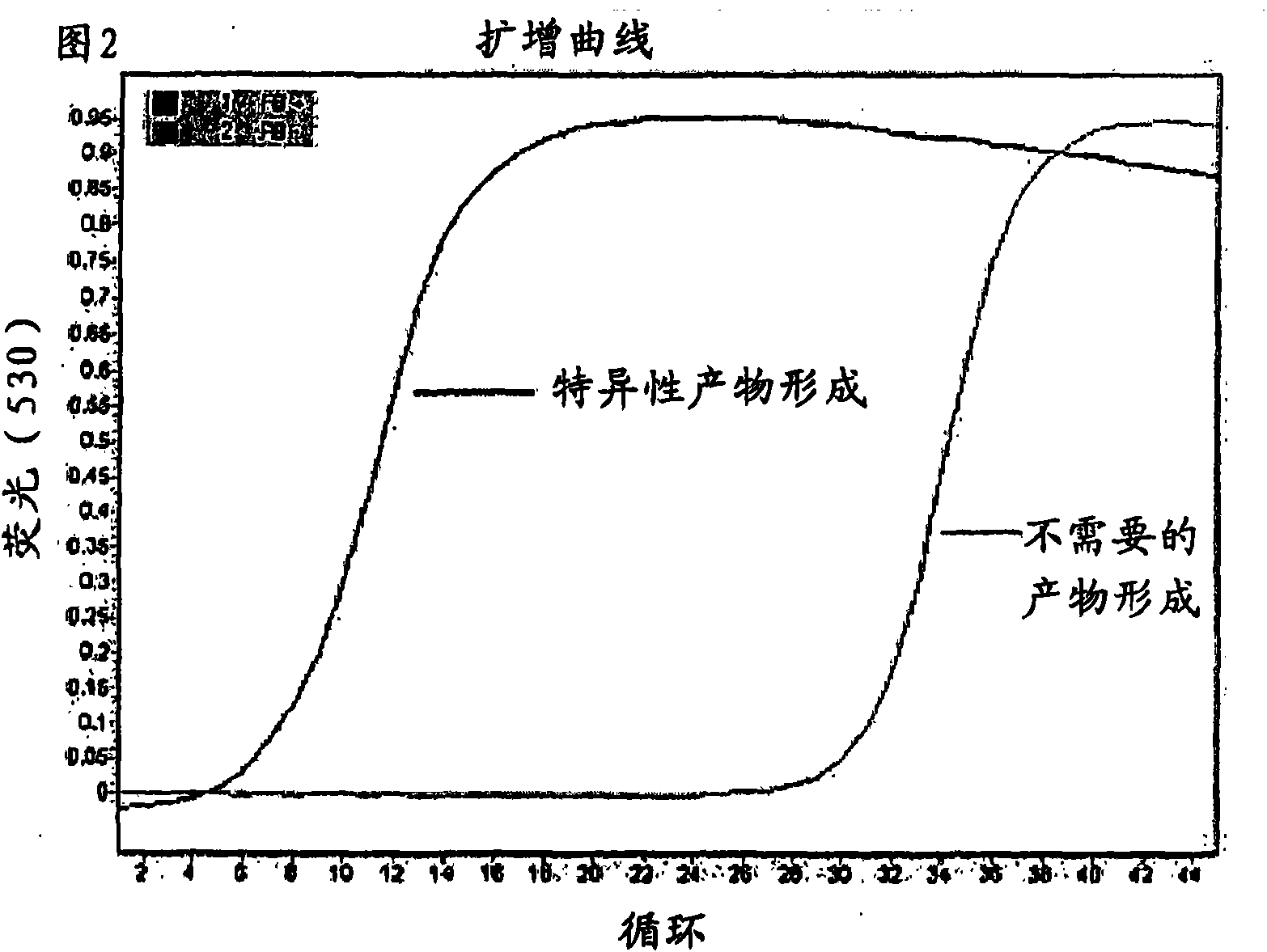
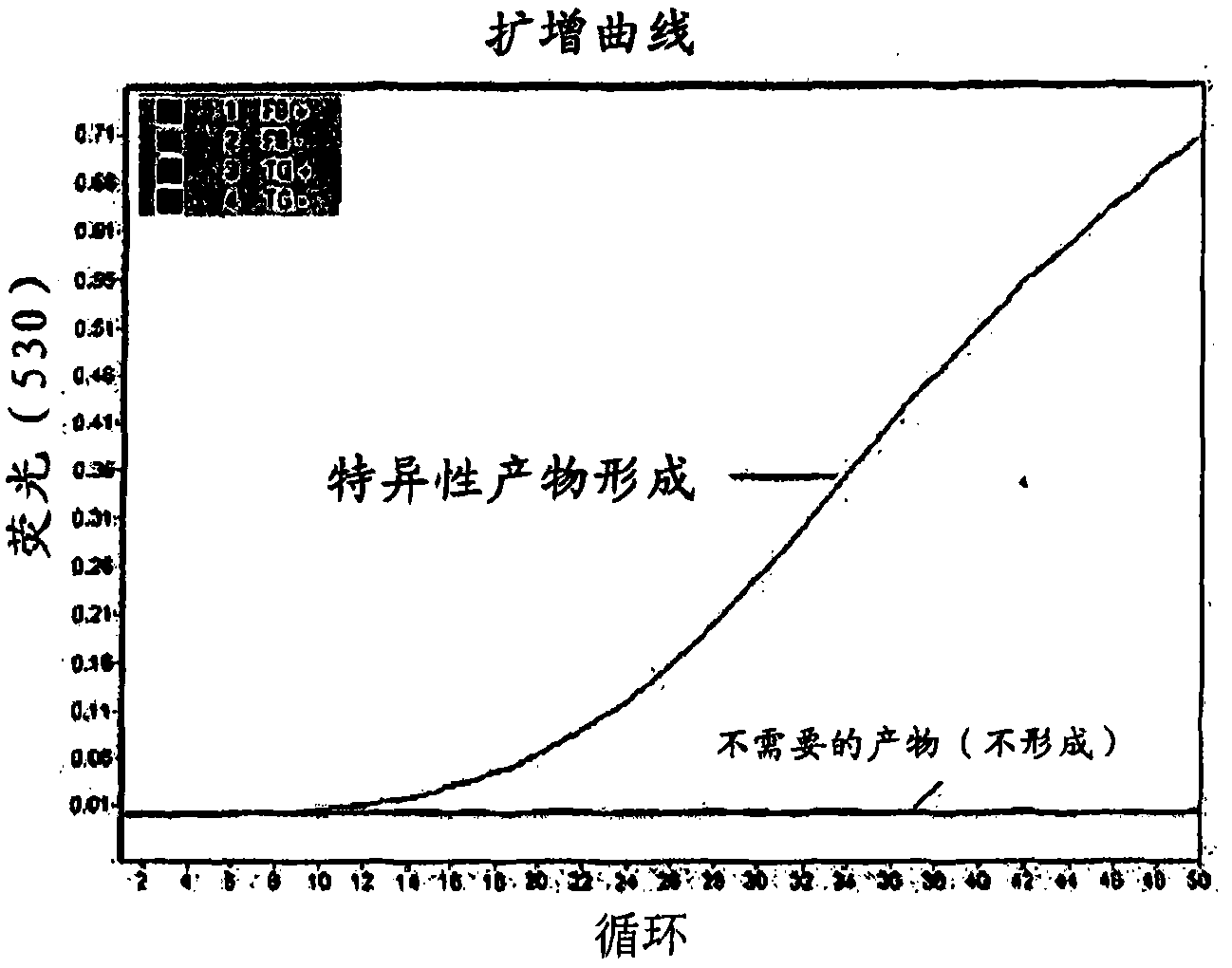
[0191] If the amount of sample DNA is insufficient to obtain amplified products from drug-sensitive regions, genome-wide enrichment can be used to amplify sample DNA before amplifying specific regions or amplicons. Genome-wide enrichment in MycoBuffer samples was performed in parallel with the same starting copy number of template DNA suspended in water only. To monitor overall M. tuberculosis genome-wide enrichment, three different target regions of the M. tuberculosis genome were selected to evaluate enrichment for each. A real-time PCR assay was used to screen for enrichment of each of these different target regions, compared to a non-enriched control sample. The point on the x-axis where the sample line starts to rise is an indication of the amount of starting genetic material in the samp...
Embodiment 2
[0314] Example 2 : Determination of drug resistance or sensitivity in human MTb samples
[0315] The purpose of these experiments was to demonstrate that clinical samples previously tested and determined to contain MTb could be rapidly assayed for drug resistance or sensitivity. Blinded clinical samples from MTb patients prepared according to the Petroff method were probed and resuspended in MGIT buffer (Becton Dickinson). The samples were assayed for rifampicin and streptomycin resistance using primer pairs, amplicons and melting temperatures as shown in Tables 2 and 3.
[0316] MTb test program:
[0317] Samples were run against the H37RV standard using the cfp32 Taqman assay to quantify the samples.
[0318] Master mix: 1x Kappa, Sybr-free buffer (Kappa Biosystems SYBRG1 master mix without SYBR), 1 μl cfp32 oligonucleotide (oligo), 1.75 mM MgCl, made up to 9 μl with water
[0319] Add 18 μl master mix per sample to 384-well plate
[0320] Add 2 μl of sample
[0321] ...
Embodiment 3
[0372] Example 3 : Determination of resistance to antifungal agents
[0373] Fungal and yeast infections cause a large number of diseases in humans. Some simple examples of clinically important fungi include:
[0374] Malassezia furfur and Exophialawneckii (surface skin)
[0375] Piedraia hortae and Trichosporonbeigelii (hair)
[0376] Microsporum species (skin and hair)
[0377] Epidermophyton species (skin and nails)
[0378] Trichophyton species (skin, hair, and nails)
[0379] Sporothrix schenckii, Cladosporium species, Phialophora species, and Fonsecaea species (subcutaneous / lymphoid tissue-chromoblastomycosis)
[0380] Histoplasma capsulatum, Coccidioidesimmitis, Fusarium species, Penicillium species (systemic respiration)
[0381] Blastomyces dermatitidis (subcutaneous / respiratory)
[0382] Cryptococcus neoformans (respiratory / CNS)
[0383] Aspergillus species, Mucor species, Candida species and Rhizopus species (opportunity to be involved in various body parts)...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com