Detection kit and detection method for 8 species of pathogenic bacteria in dairy products
A detection kit and detection method technology are applied to the detection kit for 8 pathogenic bacteria in dairy products and the detection field thereof, which can solve the problems of slow growth, low detection rate, and high requirements on culture conditions, and save labor. and financial resources, the detection time is short, and the operation is simple.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Embodiment 1, the establishment of 8 kinds of pathogenic bacteria detection kits and detection methods in dairy products
[0049] (1) Design and synthesis of primers and assembly of kits:
[0050]
[0051]
[0052] On this basis, a kit for the detection of 8 kinds of pathogenic bacteria in dairy products was designed, which included two sets of detection solvents: detection solution A and detection solution B;
[0053] Among them, detection solution A contains 10mM Tris·Cl, 50mM KCl, 25mM MgCl 2 , dNTP mixture (including 2.5 mM each of dATP, dGTP, dCTP and dTTP), 5 U / μL of Taq DNA polymerase, and 10 μM each of primer pairs of Enterobacter sakazakii, Serratia marcescens, Pseudomonas aeruginosa and Proteus vulgaris;
[0054] Detection solution B contains 10mM Tris Cl, 50mM KCl, 25mM MgCl 2 , dNTP mixture (including dATP, dGTP, dCTP and dTTP each 2.5mM), Taq DNA polymerase 5U / μL and pathogenic Aeromonas hydrophila, Proteus mirabilis, Klebsiella pneumoniae and Serra...
Embodiment 2
[0078] Embodiment 2, comparison with conventional PCR-electrophoresis detection method
[0079] The detection kit and detection method established in Example 1 involve two groups of PCR systems. The first group of composite PCR systems detects Enterobacter sakazakii, Serratia marcescens, Pseudomonas aeruginosa and Proteus vulgaris at one time; The PCR system detects pathogenic Aeromonas hydrophila, Proteus mirabilis, Klebsiella pneumoniae and Serratia odorans at one time.
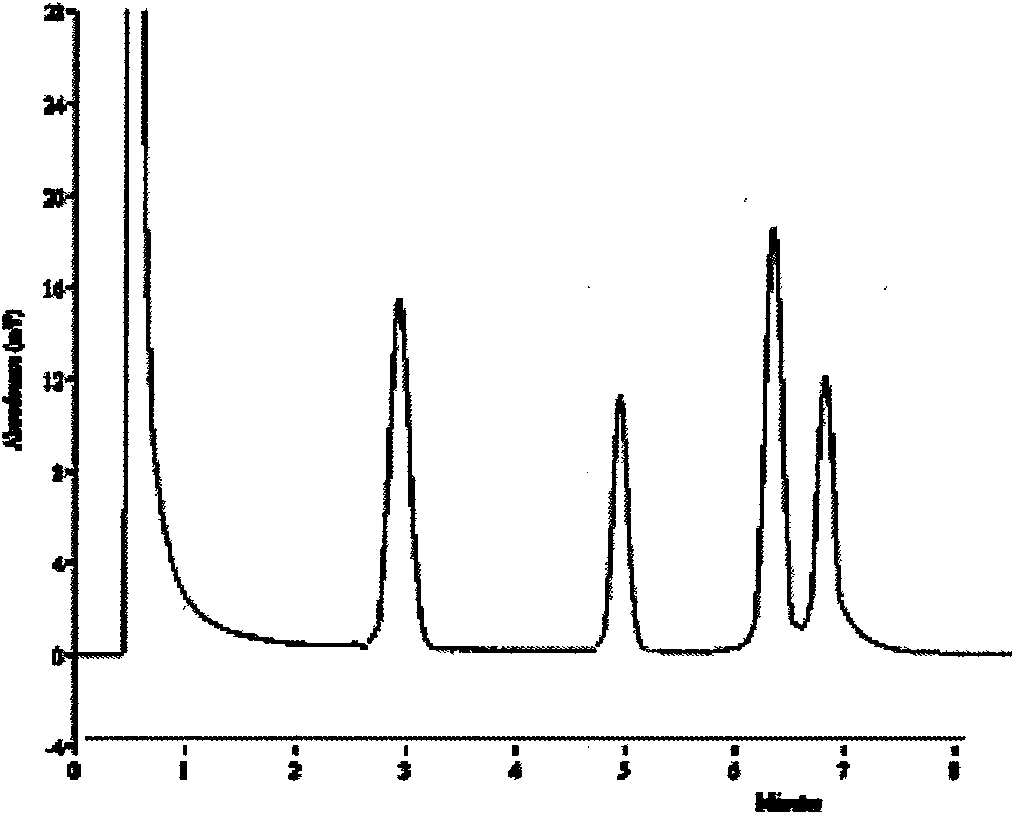
[0080] According to the method established in Example 1, the DNA of the standard bacterial strain was extracted as a template, and amplified according to the PCR conditions of the method established in Example 1: the first group of Enterobacter sakazakii, Serratia marcescens, Pseudomonas aeruginosa and The expected amplified fragments of the four kinds of Proteus vulgaris are: 228bp, 332bp, 396bp, 413bp; the second group of pathogenic Aeromonas hydrophila, Proteus mirabilis, Klebsiella pneumoniae and Serrat...
Embodiment 3
[0083] Embodiment 3, PCR-DHPLC detection method specificity test
[0084] Take the test strains listed in Table 1, extract the genomic DNA after culture, and establish the template library. Then, using these genomic DNAs as templates, the method established in Example 1 was used for PCR-DHPLC analysis and detection.
[0085] Table 1
[0086]
[0087]
[0088]
[0089]The results of the specificity test show that the detection kit and detection method established in Example 1 can specifically detect 8 kinds of target pathogenic bacteria after two detections; Serratia aeruginosa, Pseudomonas aeruginosa and Proteus vulgaris detected specific sample peaks; detection using detection reagent B, only pathogenic Aeromonas hydrophila, Proteus mirabilis, Klebsiella pneumoniae and odor Serratia detected a specific sample peak, and the detection results of other control bacteria were all negative, without false positive and false negative results, and had high specificity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com