Real-time quantitative fluorescence PCR test method based on double external references of RNA and DNA and application thereof
A real-time quantitative fluorescence and detection method technology, applied in the field of bioinformatics, can solve problems such as no copy number linking, and achieve the effect of improving experimental design
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] 1. Treatment of experimental silkworms:
[0041] The eggs of the experimental silkworm 54A were provided by the Sericulture Research Institute of the Chinese Academy of Agricultural Sciences, and were reared with artificial feed at 25°C after being bred and hatched. The larvae were dissected on the 3rd day when they reached the 5th instar, and the middle silk gland, posterior silk gland, fat body, Malpighian duct and midgut were collected, washed with redistilled water three times, and stored at -70°C.
[0042] 2. Selection of RNA and DNA external reference:
[0043] The piggyBac transposon was found in the Lepidoptera cell line TN-368, which consists of two transposable arms and a coding sequence for the transposase IFP2. Since IFP2 does not exist in silkworm, IFP2 was selected as the external reference in this embodiment.
[0044] 3. Detect the ratio of RNA and DNA external reference:
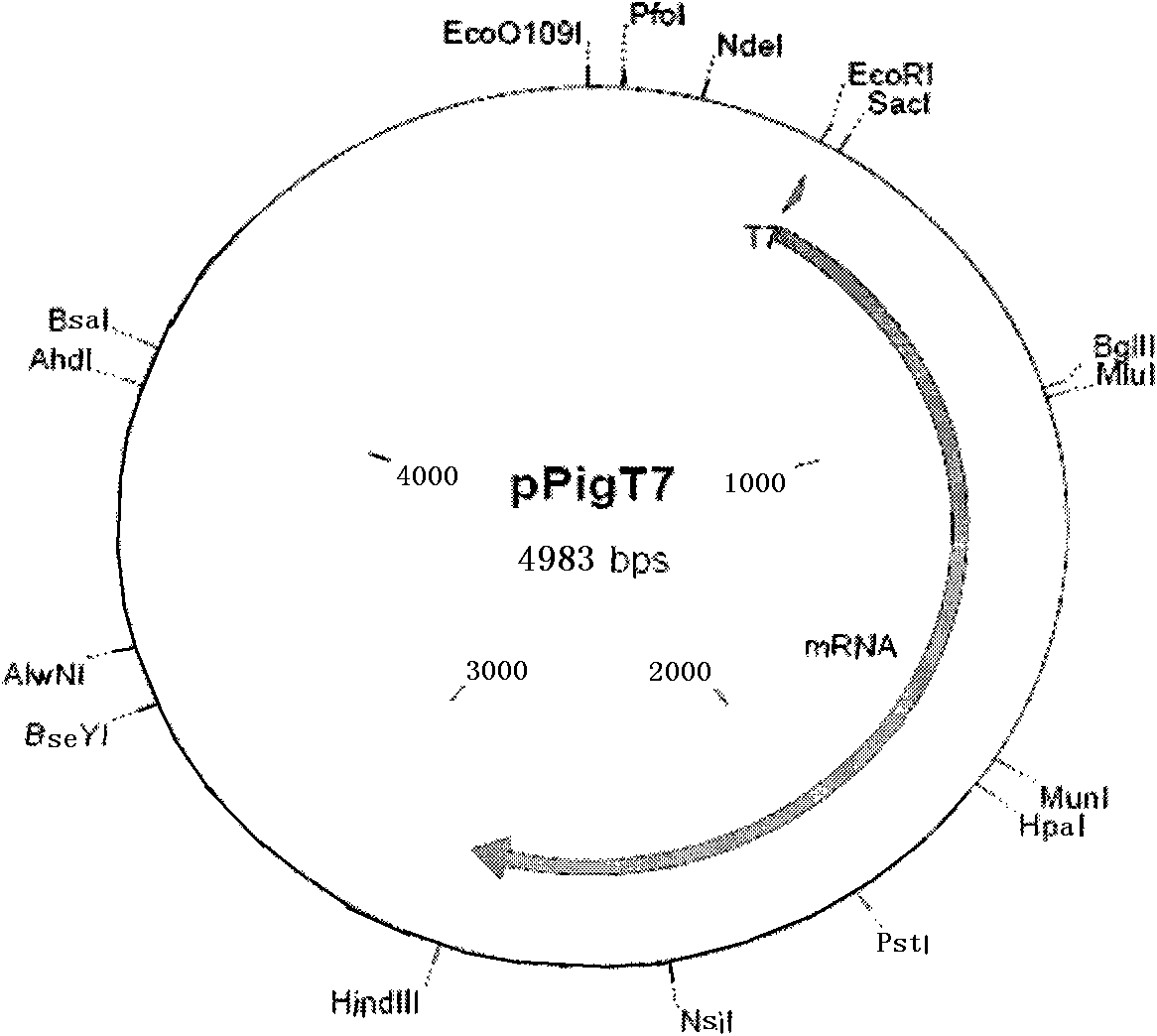
[0045] Plasmid pPigT.7 is made by inserting T7-IFP2 expression cassette into ve...
Embodiment 2
[0108] Example 2 Detection of A3, GAPDH, and 28S rRNA expression in various tissues of the silkworm:
[0109] Use the detection method in Example 1 to A3, GAPDH, the transcription product of 28S rDNA gene and gene and external reference IFP2mRNA and DNA in silk gland of Bombyx mori middle part, posterior silk gland, fat body, Malpighian tubule and midgut extract The copy number in was determined. A3, the expression levels of GAPDH and 28SrDNA genes per copy in these tissues were calculated according to the above method. Their apparent transcript levels vary greatly among different tissues (as shown in Table 3).
[0110] Table 3 Apparent transcription levels of A3, GAPDH and 28S rRNA genes in silkworm tissues
[0111] organize
A3
GAPDH
28S rRNA
middle silk gland
(MSG)
122.25±4.83
10.794±1.728
5935.5±353.3
posterior silk gland
(PSG)
1097.6±276.4
154.39±6.72
9016.1±458.8
...
Embodiment 3
[0115] The detection of the expression of silk fibroin gene and sericin 1 gene in embodiment 3 silkworm:
[0116] The detection method is the same as in Example 1. The apparent transcription levels of silk fibroin heavy chain gene (FibH), light chain gene and sericin 1 gene in each tissue of silkworm were determined. The results showed that FibH and FibL were exclusively expressed in the posterior silk gland, and Ser-1 was exclusively expressed in the middle silk gland. The apparent transcription level of FibL in the posterior silk gland (192029±30739) was similar to that of Ser-1 in the middle silk gland (196539±12455), while the apparent transcription level of FibH in the posterior silk gland (106453 ±13667) is about half of FibL.
[0117] If the expression of FibH and FibL in the posterior silk gland and the expression of Ser-1 in the middle silk gland were normalized with A3, GAPDH or 28S rRNA as usual, the expression of Ser-1 in the middle silk gland was higher than tha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com