Method for preparing live bacteria internal standard based on gene substitution technique
A technology of gene replacement and live bacteria, which is applied in the field of preparation of live bacteria internal standards, to improve accuracy and avoid false negative effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
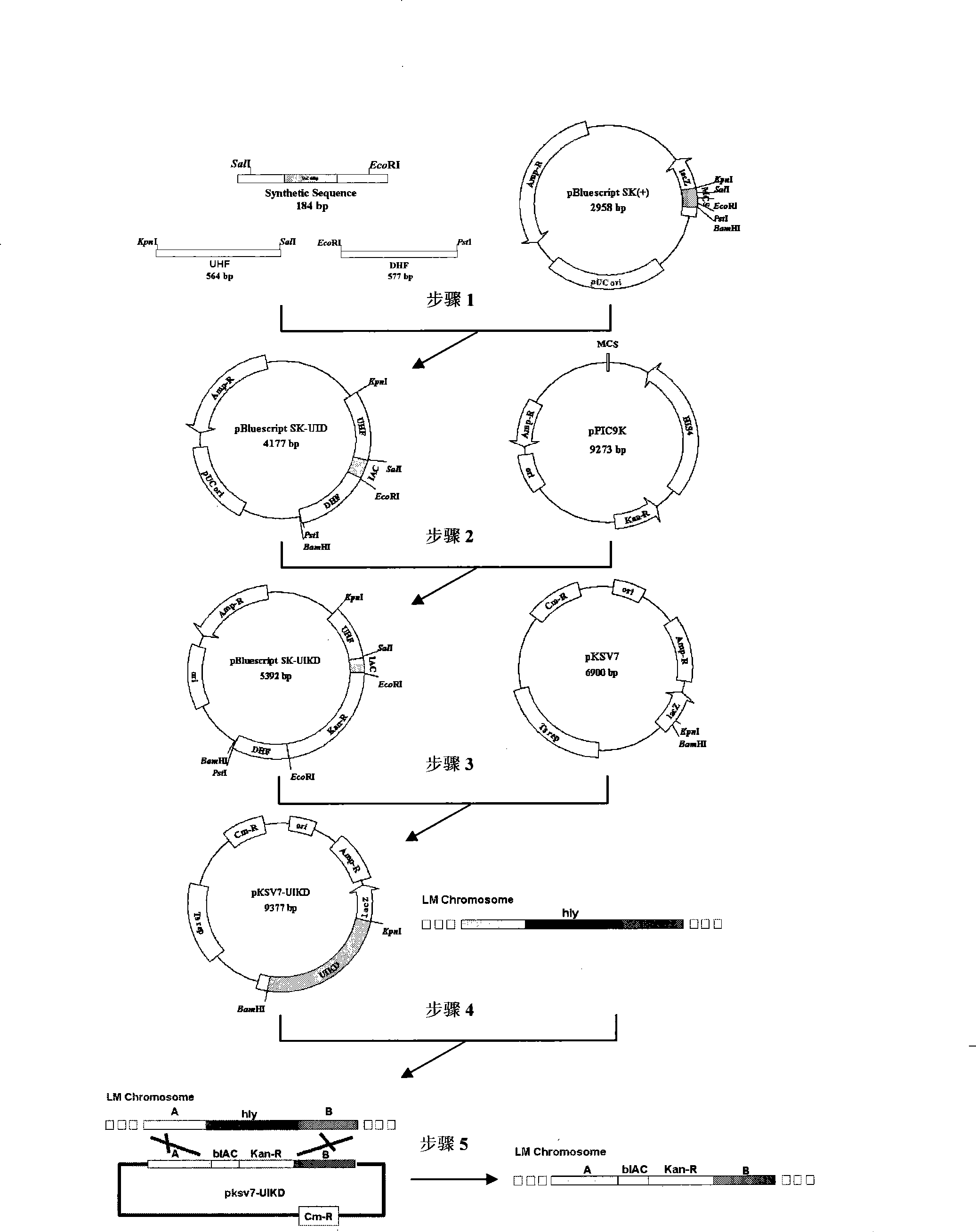
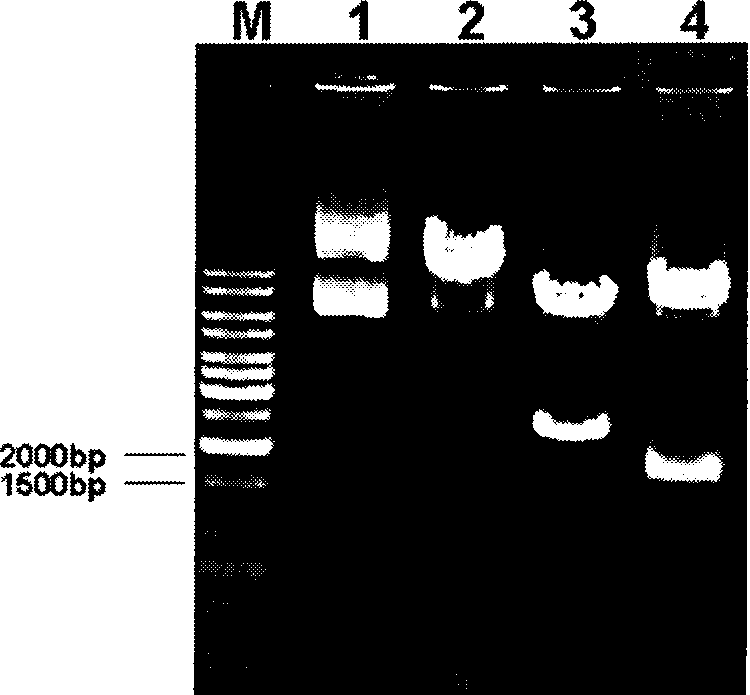
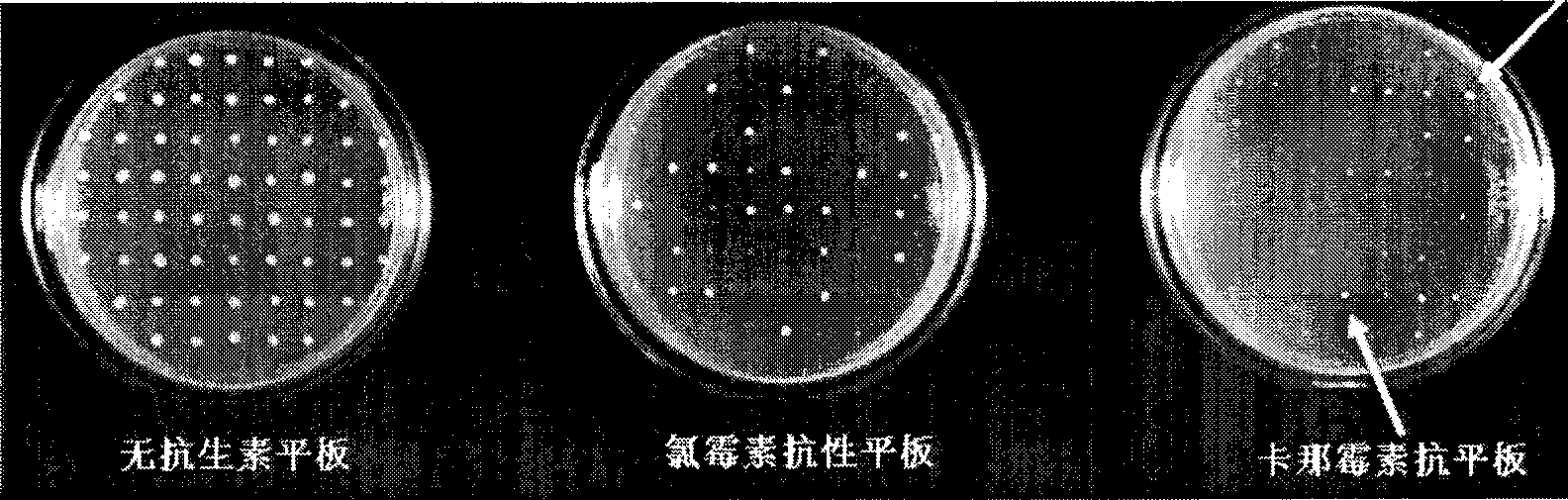
[0032] 1. Design of homologous recombination fragments and corresponding enzyme cutting sites
[0033] 1. Selection of upstream homologous sequence (UHF) and design of restriction site
[0034] According to the hly gene and its upstream fragment sequence characteristics of Listeria monocytogenes, the upstream homologous sequence amplification primers and corresponding enzyme cutting sites (Kpn I, Sal I) were designed:
[0035] Amplification primers for upstream homologous sequences (Hlyku-s / Hlyku-a)
[0036] Hlyku-s: 5'- CTTGGTACCCGATGTACCGTATTCCCTG -3'
[0037] Hlyku-a: 5'- ATTGTCGACGGGTTTCACTCTCCTTCT -3'
[0038] (a) Sequence characteristics of upstream homologous sequences:
[0039] * Length: 551 bp
[0040] *Type: nucleic acid
[0041] * Chain type: double chain
[0042] *Topology: Linear
[0043] (b) Molecule type: DNA
[0044] (c) Original source: Listeria monocytogenes
[0045] (d) Sequence description: SEQ ID NO.1
[0046] CGATGTACCGTATTCCTGCTTCTAGTTGTTGG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com