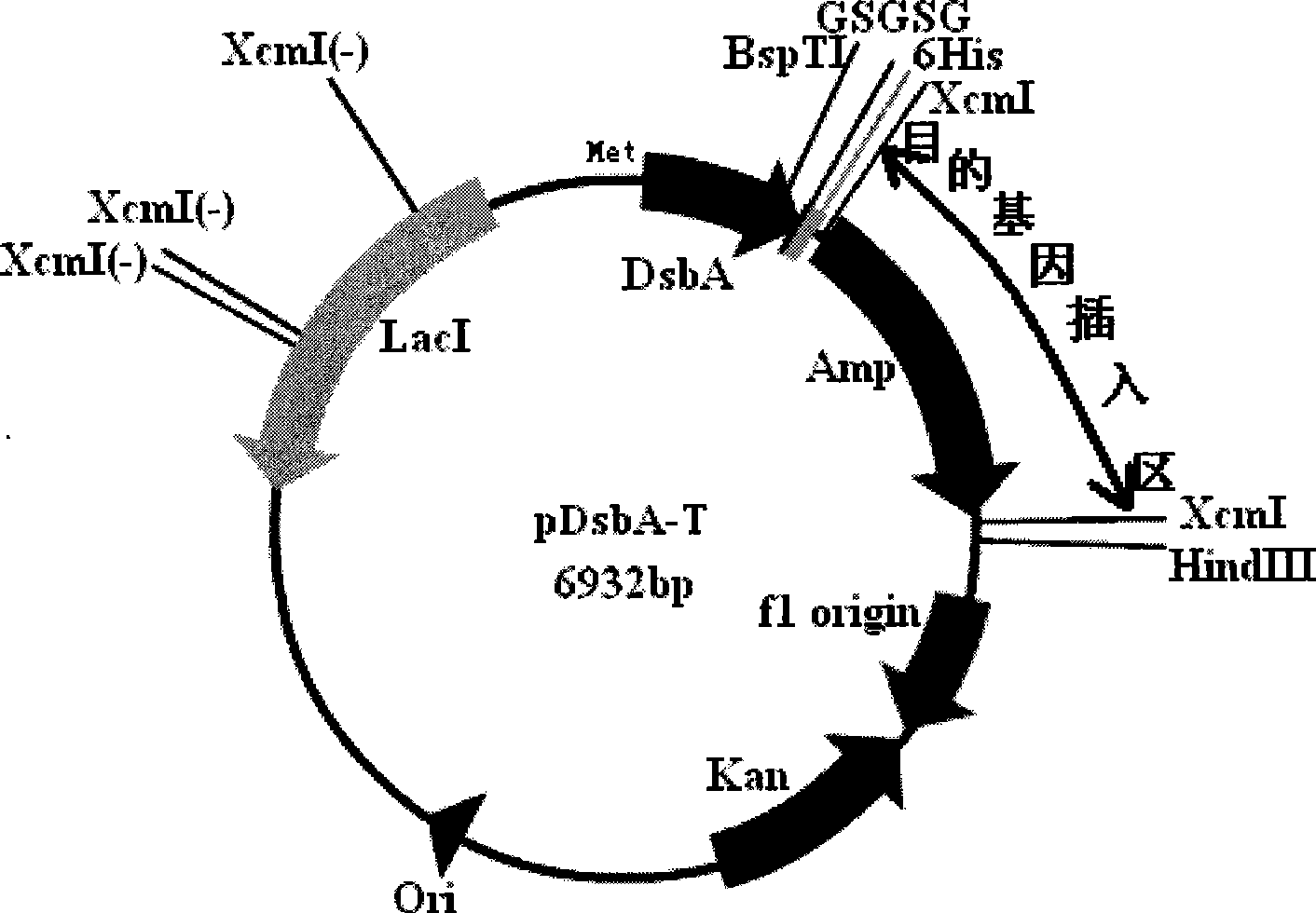
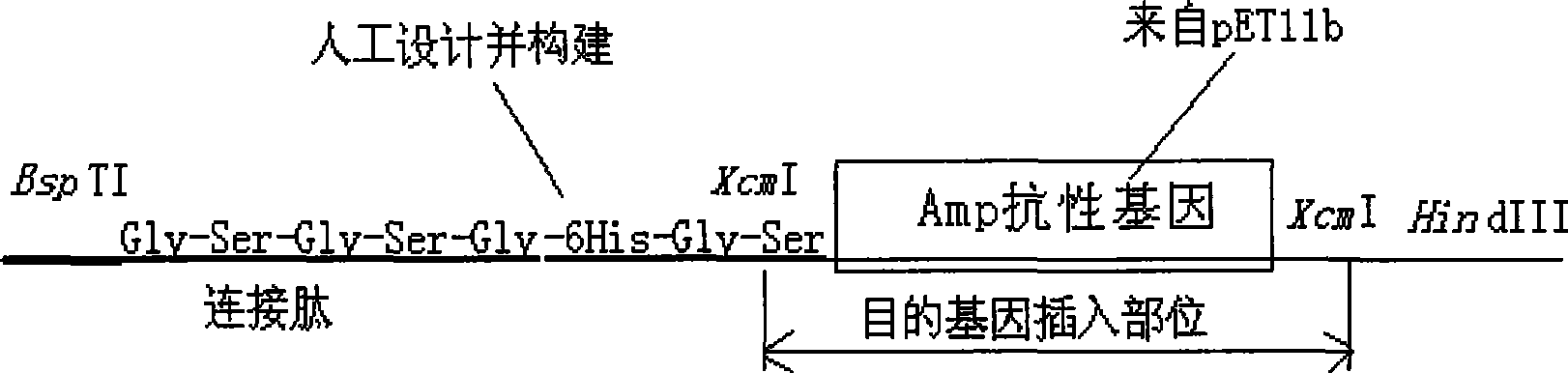
Intracellular fusion expression type pre-T vector, preparation and application
A technology of internal fusion and vector, which is applied in the field of intracellular fusion expression type pre-T vector, can solve the problems of negative cloning and connection, and achieve the effect of rapid construction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Embodiment 1: LacI gene site-directed mutation eliminates XcmI site
[0053] Using the SOE method, using pET39 as a PCR template, the six primers were designed as follows:
[0054] Lac-1: 5'-ttaggatccgcgacccatttgct-3' (including BamHI site)
[0055] Lac-2: 5'-agagatatccgcaccaacgcgca-3' (including EcoRV site)
[0056] Lac-3: 5'-ctgattggcgttgctacctccagtctggccctgca-3' (for mutation site 1)
[0057] Lac-4: 5'-gccagactggaggtagcaacgccaatcagcaacga-3' (for mutation site 1)
[0058] Lac-5: 5'-tcccactgcgatgttagttgctaacgatcagatggcgct-3' (for mutation sites 2 and 3)
[0059] Lac-6:5'-catctgatcgttagcaactaacatcgcagtgggaacgatgc3' (for mutation sites 2 and 3)
[0060] Table 2: Sequence comparison before and after site mutation
[0061] Original sequence (with XcmI recognition site) New sequence (no XcmI recognition site) bit
point
1 Gccacctccagtctggcc
GCC ACC TCC AGT CTG GCC
Ala Thr Ser Ser Leu Ala
gcTacctccagtctggcc
GCT ACC TCC AG...
Embodiment 2
[0074] Embodiment 2: the preparation of XcmI restriction cassette and the construction of front T carrier
[0075] Primers A and B serve as templates for PCR1 reaction:
[0076] A: 5′-ATACTTAAGCGAGAAAAAAGGTTCTGGTTCTGGTCAT-3′
[0077] B: 5′-GACATTAACCTATCCATTAGATCCATGGTGATGATGATGATGACCAGAACCAGAACC-3′
[0078] The reaction parameters were as follows: denaturation at 94°C for 10 min, annealing at 48°C for 300 s, and extension at 72°C for 10 min.
[0079] With pET11b as template, utilize following two primers to carry out PCR2 reaction:
[0080] Amp1: 5′-CACCATGGATCTAATGGATAGGTTAATGTCATGATAATA-3′
[0081] Amp2:5′-CTTAAGCTTCCAAGGGTATAATGGAAATGAAGTTTTAAATCAATC-3′
[0082] The reaction parameters were as follows: pre-denaturation at 94°C for 3 min, denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 120 s, 30 cycles, and extension at 72°C for 10 min.
[0083]The above two PCR1 and PCR2 product fragments were used for gene splicing reaction with prime...
Embodiment 3
[0085] Embodiment 3: the removal of DsbA signal peptide gene (keep ATG as initiation codon):
[0086] Design primers:
[0087] DsbA-1:5'-AAG CATATG GCGCAGTATGAAGAT-3', (the underlined part is NdeI recognition sequence)
[0088] DsbA-2:5'-TCG CTTAAG TATTTCACTGT-3', (the underlined part is the recognition sequence of BspTI)
[0089] PCR template: pET39 plasmid (or Escherichia coli genomic DNA is also available, pET39 plasmid is recommended as a template)
[0090] PCR parameters: 94°C for 3min
[0091] 94℃ 30s 40℃ 30s 72℃ 1min 3 cycles
[0092] 94℃ 30s 48℃ 30s 72℃ 1min 27 cycles
[0093] 72°C 10min
[0094] 4°C 10min
[0095] Digest the PCR product and pET39-2 with NdeI and BspTI, replace the original DsbA gene in pET39-2 with the DNA fragment without the signal peptide gene, and obtain a recombinant plasmid without the DsbA signal peptide gene, that is, the intracellular fusion expression type T vector creates conditions for realizing t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com