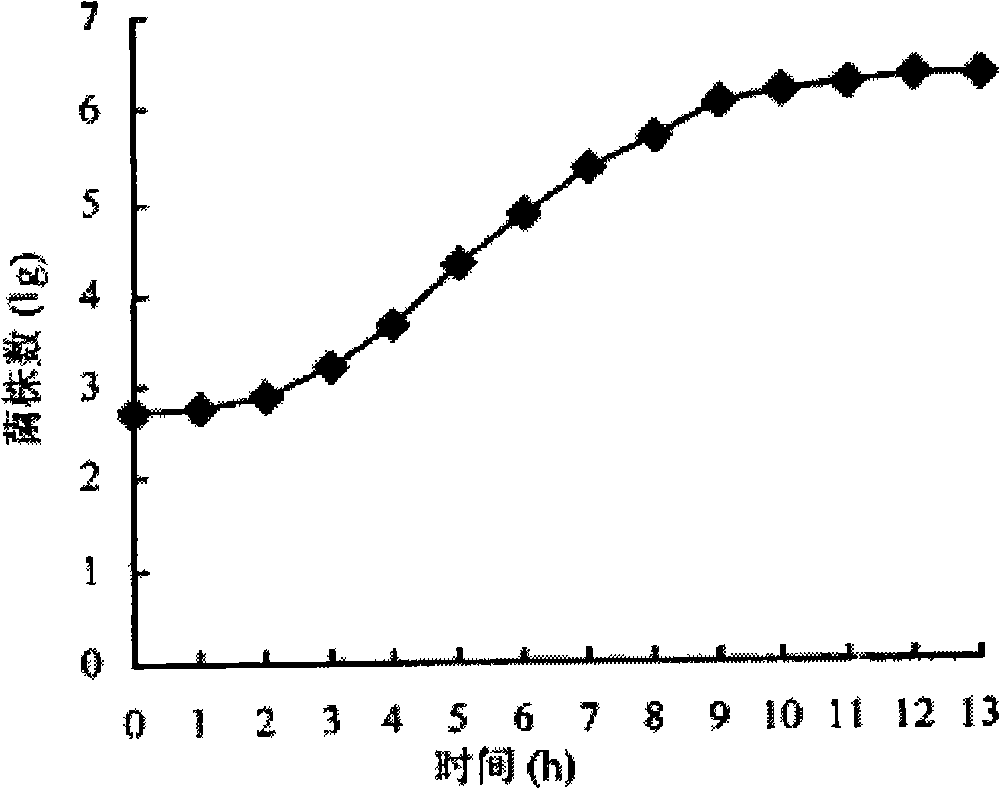
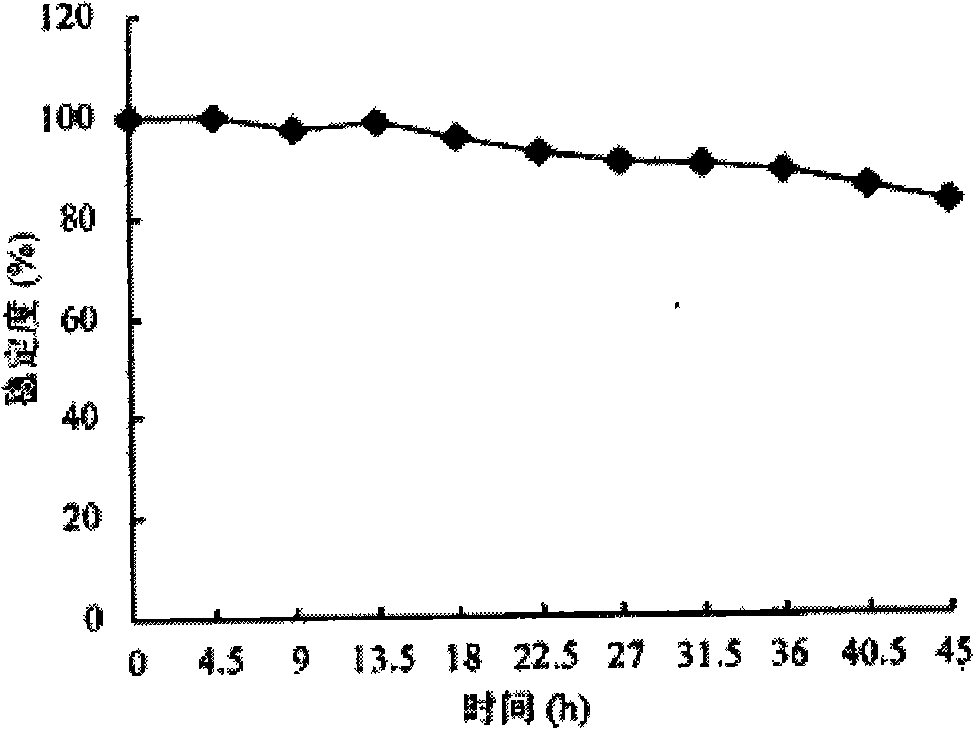
Construction method of onion pseudomonas genetic engineering bacteria
A technology of Pseudomonas cepacia and genetically engineered bacteria, applied in the field of genetic engineering, can solve problems such as low efficiency and yield, inactive heterologous expression of Pseudomonas cepacia lipase, and unclear metabolic regulation mechanism, etc., to achieve Good low-carbon alcohol tolerance and high temperature resistance, high expression and catalytic activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0017] Example 1: Construction of lipase genetically engineered bacteria.
[0018] 1. Construction of Homologous Recombination Engineering Bacteria
[0019] 1) Cloning of related genes
[0020] PCR method was used to amplify T7 RNA polymerase gene, lipase gene upstream segment (homologous recombination segment gene 1) and lipase gene downstream segment (homologous recombination segment gene 2), Tmp resistance gene, lipase and its partner gene.
[0021] Primers for T7 RNA polymerase gene
[0022] Upstream primer 5': CTT CTGCAG ATGAACACGATTAACATCGCT
[0023] Downstream primer 5': CTTCTGCAG TTACGCGAACGCGAAGTCCGA
[0024] Primer for homologous recombination segment gene 1
[0025] Upstream primer 5': CTTGCGGCCGC AGCATCGCTACGCGCTGAAC
[0026] Downstream primer 5': CTTCTGCAG GCATGTTCTCCTGATTATTG
[0027] Primer for homologous recombination segment gene 2
[0028] Upstream primer 5': CTTCTGCAG AGATGTTGCTCGATGGTG
[0029] Downstream primer 5': CTTCTCGAG GTGATCTACGTCGGCAGTCT ...
example 2
[0070] Example 2: Recombinant bacteria are suitable for the expression of the α-amylase gene regulated by the T7 promoter
[0071] 1. Acquisition of α-amylase gene
[0072] Primer
[0073] Upstream primer 5': AAAGGATCC ACC ATC CTT CAT GCC TGGA
[0074] Downstream primer 5': CTTAAGCTT TTA ATG AGG AAG AGA ACC CGCT
[0075] Using Bacillus subtilis XL-15 genome DNA as a template, the α-amylase gene was amplified by PCR. An XbaI restriction site was added to the upstream primer, a HindIII restriction site was added to the downstream primer, and the PCR amplification product was simultaneously digested with XbaI and HindIII and cloned into the expression vector pBBR22b to obtain pBBR22AMY.
[0076] 2. α-amylase expression vector transforms Pseudomonas cepacia.
[0077] The pBBR22AMY expression plasmid was introduced into the constructed Pseudomonas cepacia strain containing T7 RNA polymerase gene to obtain α-amylase expression engineering bacteria. The detailed ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com