Method for preparing firefly luciferase
The technology of luciferase and luciferase gene is applied in the field of preparing firefly luciferase, which can solve the problems of high breeding cost, long production cycle, complex enzyme composition and the like, and achieve the effects of easy purification, low cost and simple process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
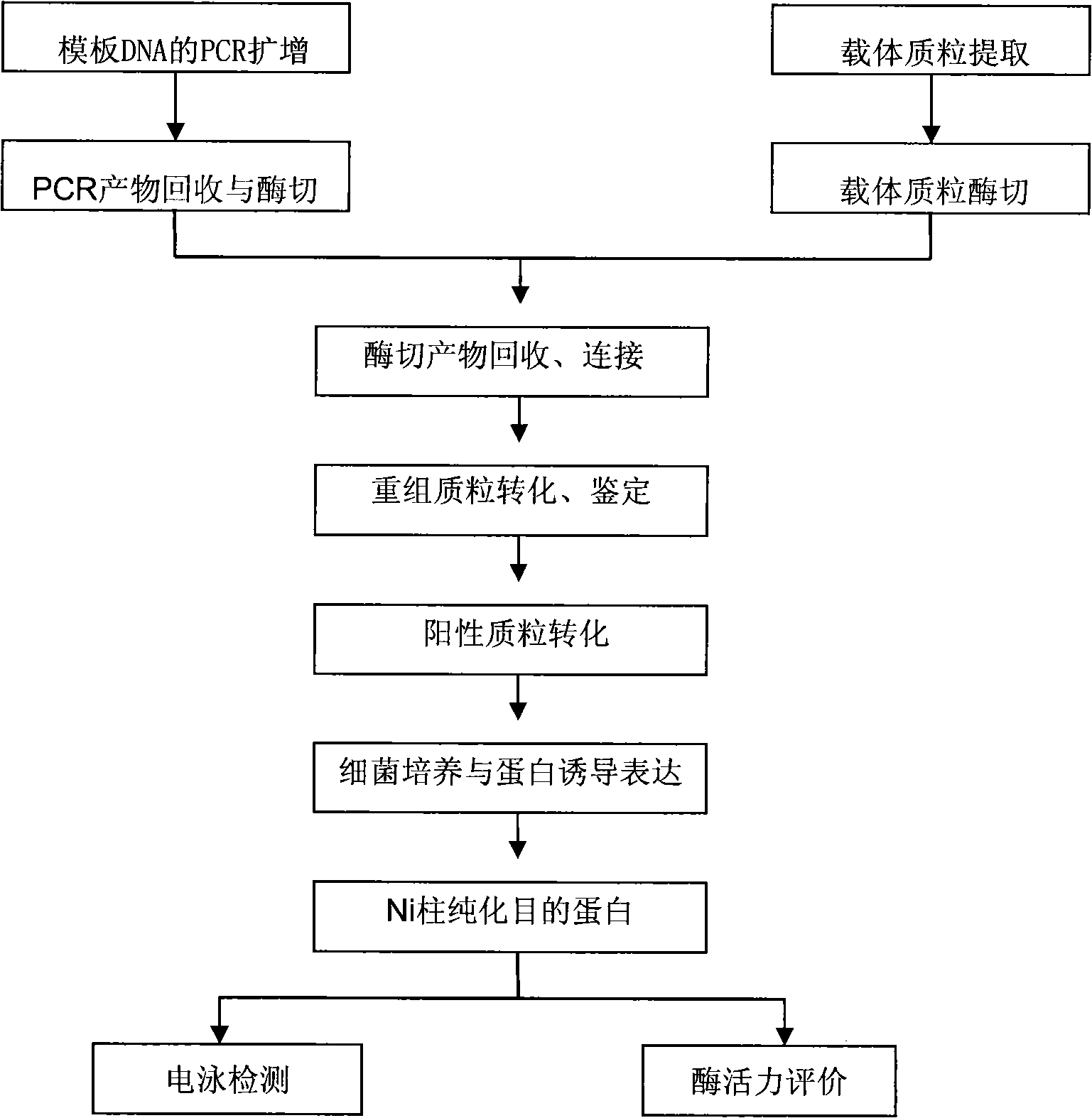
[0020] Embodiment 1, the construction of the recombinant expression vector containing firefly luciferase gene (luc)
[0021] 1. Primer design
[0022] According to the nucleotide sequence of the vector pGEM-luc (purchased from Promega) containing the North American firefly (P. The multiple cloning site of pET28a and the restriction endonuclease site of luc gene were used to design primers. The NdeI restriction site was introduced at the 5' end of the upstream primer, and the XhooI restriction site was introduced at the 5' end of the downstream primer. The primers were synthesized by Shanghai Sangong Company. The specific sequences are as follows:
[0023] Upstream primer luc-R: 5'-GGCCGG CATATG GAAGACGCCAAAAAC-3'
[0024] Downstream primer luc-F: 5'-GGCC CTCGAG TTACAATTTGGACTTTCC-3'
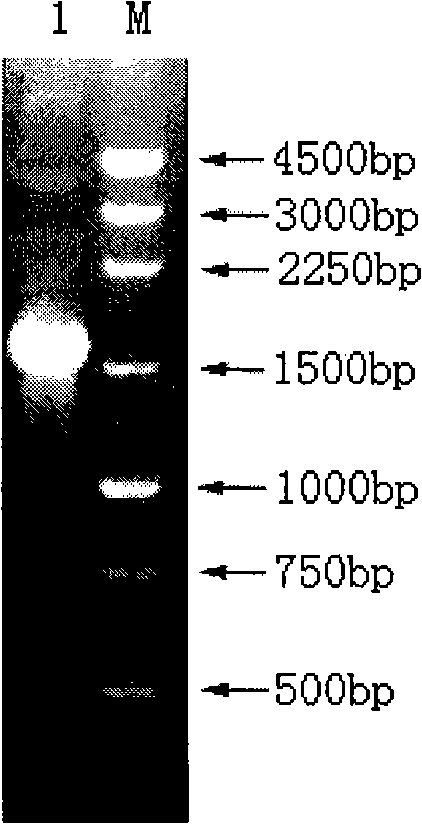
[0025] 2. PCR amplification of luciferase gene (luc)
[0026] Using the vector pGEM-luc (purchased from Promega) containing the luc gene as a template, PCR amplification was carried out, ...
Embodiment 2
[0046] Embodiment 2, induced expression of luciferase
[0047]The recombinant expression vector pET28a-luc of the above-mentioned embodiment 1 is transferred in the competent cell of Escherichia coli BL21 (DE3); Pick a single colony and inoculate it in 2 ml of LB culture fluid containing kanamycin with a final concentration of 50 μg / ml , 37 ° C 200 rpm / min shaking culture overnight. Expand the culture of the bacterial suspension at a volume ratio of 1:100, when the OD of the bacterial liquid 600 When the value reaches 0.8, take 0.5ml of bacterial solution in different concentrations of isopropyl-β-D-thiogalactoside (IPTG) (0.5, 1, 1.5, 2mmol / L), different temperatures (22 ° C, 37 ° C ) and at different times (2, 4, 6, 12 and 18h) to optimize the induction conditions. At the same time, Escherichia coli BL21 (DE3) transfected with pET28a and not induced with IPTG were used as controls.
[0048] Take the Escherichia coli BL21(DE3) transformed into the recombinant expression ve...
Embodiment 3
[0050] Embodiment 3, separation and purification of luciferase
[0051] Since the recombinant expression vector pET28a-luc constructed in the above-mentioned Example 1 has a fusion tag (His·Tag) gene sequence, the expressed target protein also has a His·Tag tag. Therefore, purification of the target protein can be performed by affinity chromatography using Ni-complexed agarose particles (90 μm). Affinity chromatography columns were purchased from GE Healthcare.
[0052] The Escherichia coli BL21(DE3) transformed into the recombinant expression vector pET28a-luc was cultured in LB medium containing kanamycin, and after the expression was induced with IPTG according to the method of the above-mentioned Example 2, it was harvested by centrifugation at 5000rpm / min for 10min Bacteria precipitation; re-dissolve the bacterial precipitation with BindingBuffer (20mM sodium phosphate, 0.5M NaCl, 20mM imidazole, pH7.4) containing a final concentration of 0.2mg / ml lysozyme, and ultrasoni...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Light intensity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com