Sphingosine kinase-1 mediates gene expression regulation of a monocyte chemoattractant protein-1 gene
A technology of sphingosine kinase and monocytes, applied in the field of gene expression regulation of monocyte chemoattractant protein-1 gene mediated by sphingosine kinase-1, can solve the problem of no small molecule antagonist
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0165] cDNA microarray study of gene expression in HMVEC
[0166] In an effort to rapidly evaluate the potential global role of SK1 in endothelial cell function, cDNA microarray analysis was performed in HMVECs in the absence and presence of the SK1 inhibitor DMS. Cells are stimulated by two receptor subtypes: G protein-coupled receptors, the protease-activated receptor (PAR) family with thrombin as an agonist; and cytokine receptors with TNF-α as an agonist.
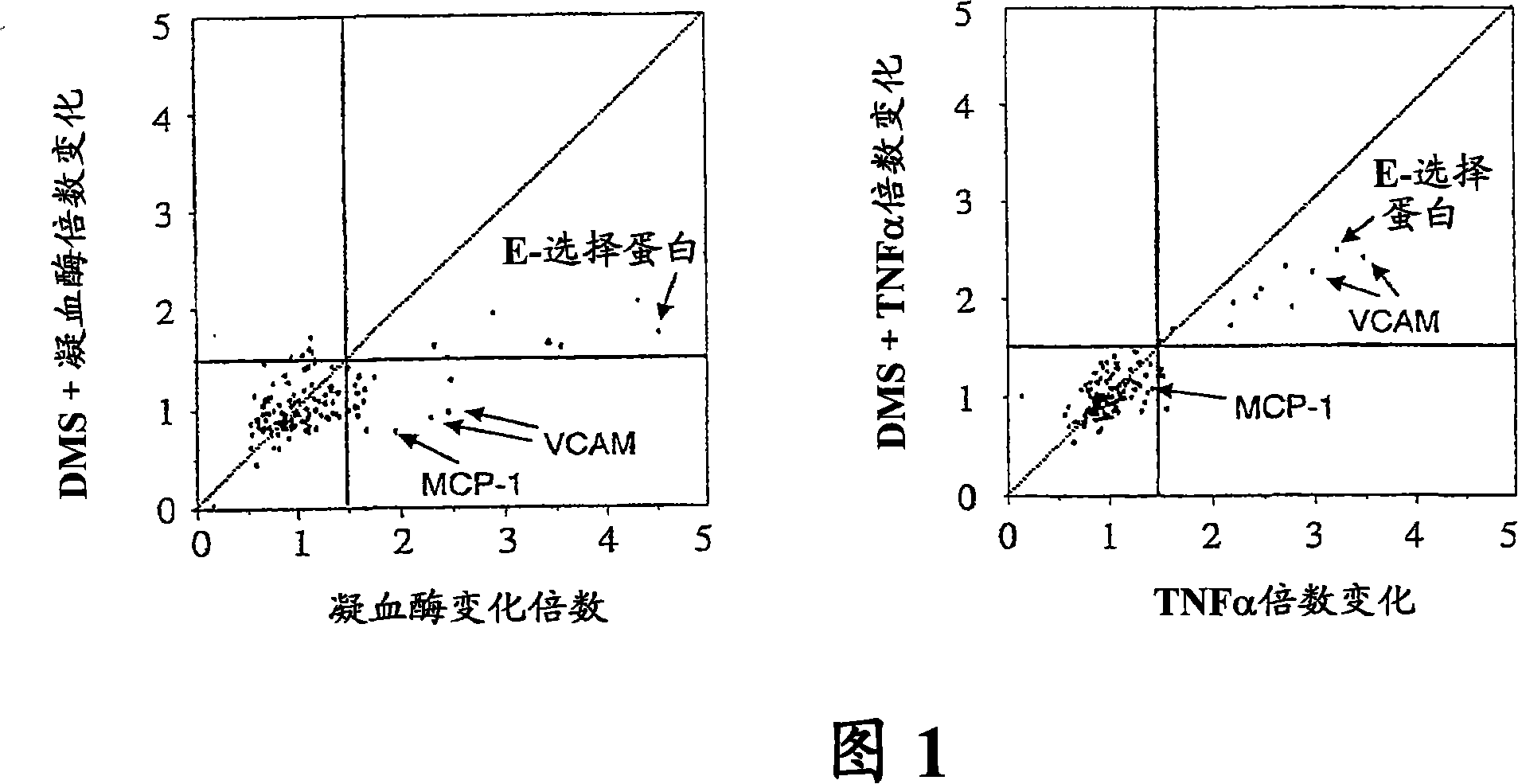
[0167] A total of 138 genes were identified that were either induced or repressed under one of the experimental conditions. Fold change results for stimulated cells without and with DMS were plotted along two axes, thus generating dot blots to visualize genes detected above a selected threshold (Figure 1). These analyzes revealed that sphingosine kinase is linked to signaling through the thrombin receptor, an event that had never been observed before. However, this is the first example of previous studies demonstratin...
Embodiment 2
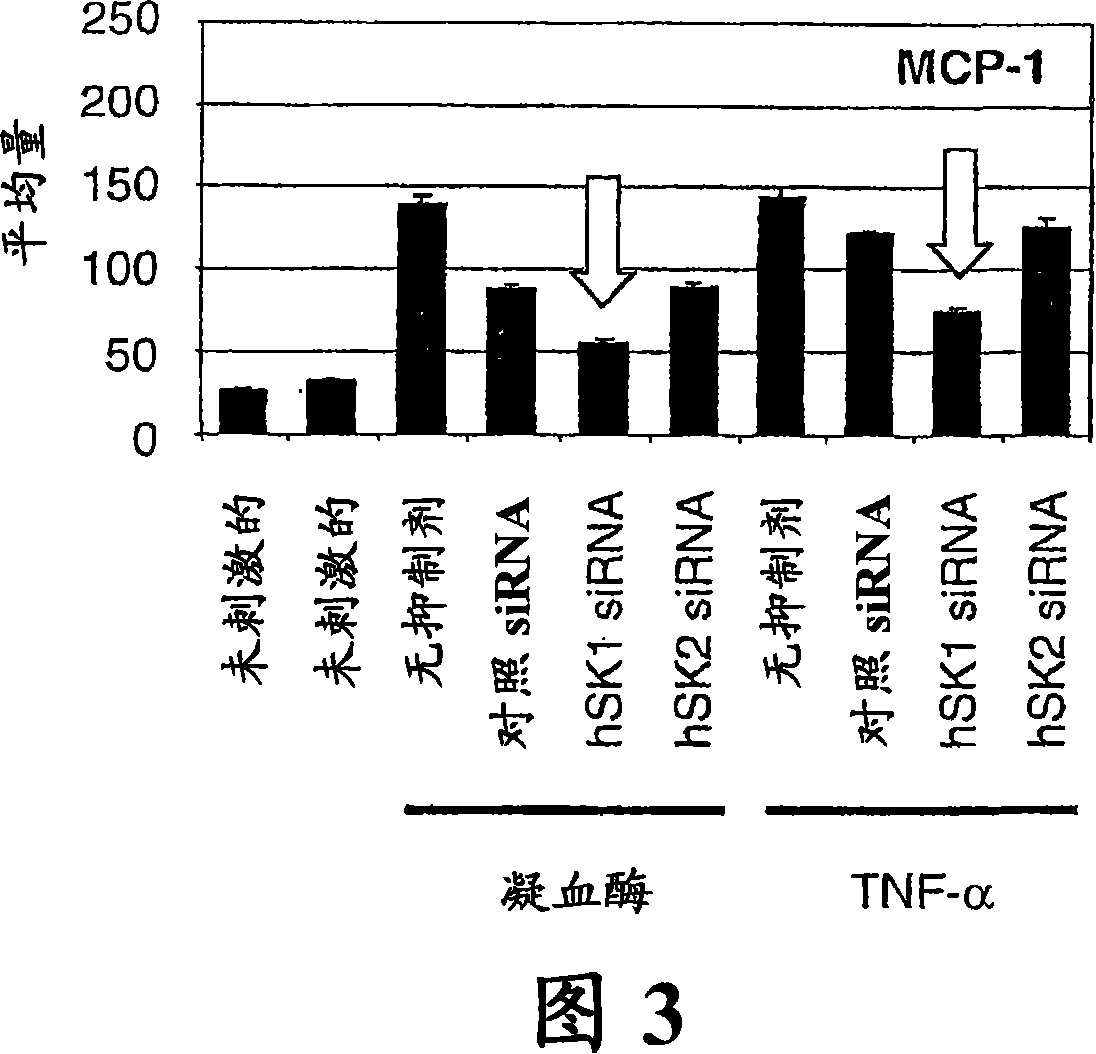
[0174] Specific inhibition of SK-1 abolishes induction of MCP-1 in HMVECs
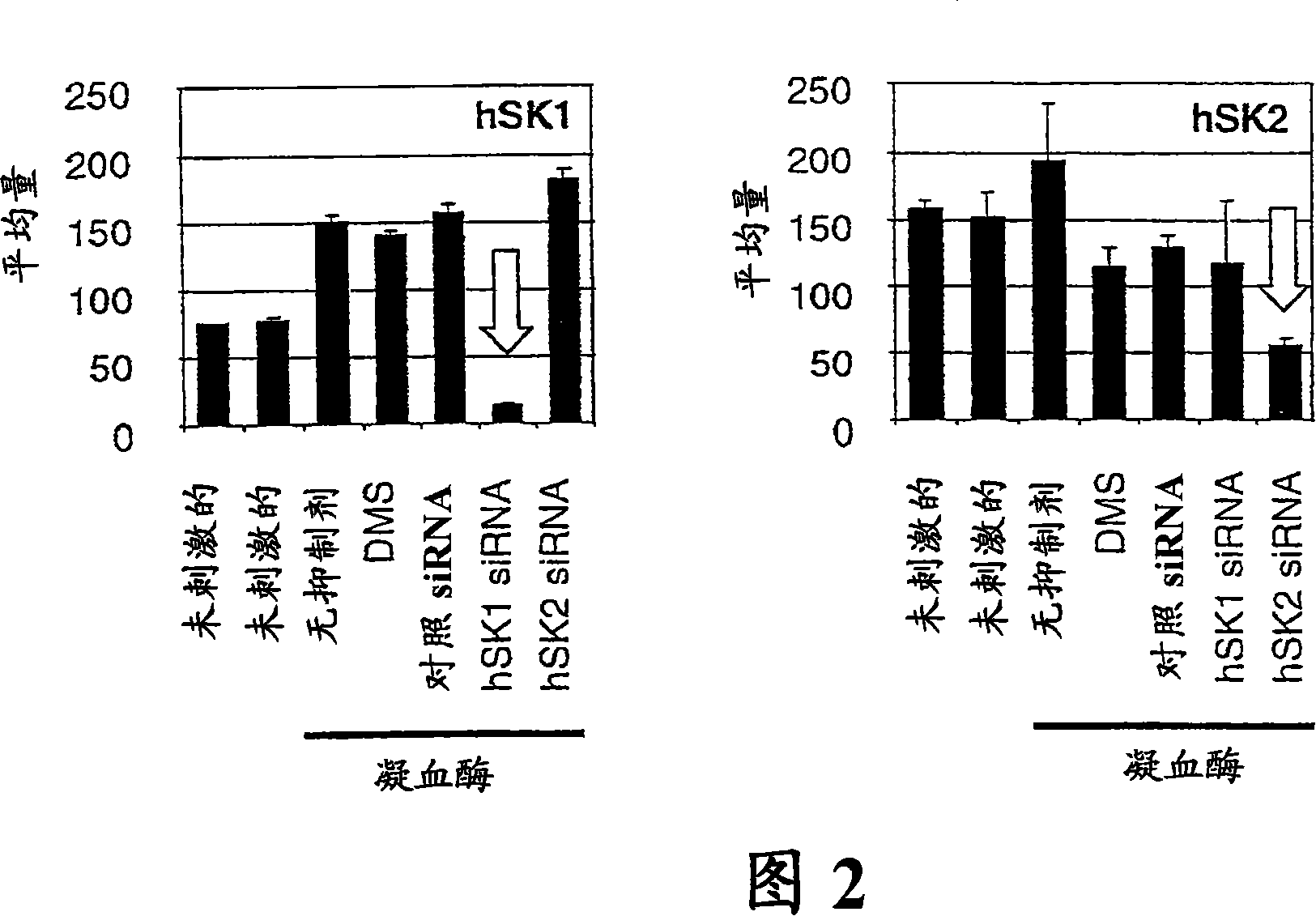
[0175] Although DMS has been shown to inhibit SK activity at lower concentrations, it can also affect the activity of protein kinase C family members as well as casein kinases at higher concentrations. Therefore, small interfering RNA (siRNA) was used to selectively inhibit SK by specifically targeting SK family members, specifically human SK1 or SK2. With the addition of fluorescein to the 3' end of the siRNA, we were able to observe nearly 100% siRNA transfection efficiency. The designed siRNAs showed specificity to their respective human SK (hSK) isoforms as detected by Taqman quantitative RT-PCR (Figure 2). When hSK1 transcripts were examined, it was observed that only hSK1-specific siRNA inhibited hSK1 expression, while hSK2-specific siRNA and control non-silencing siRNA had no effect on hSK1 expression pattern. Similarly, when detecting hSK2 transcript levels, only hSK2-specific siRNA affected ...
Embodiment 3
[0180] PAR expression in HMVEC is restricted by PAR-1, PAR-2 and PAR-4
[0181] Thrombin, a serine protease, can act on many substrates, but we were interested in PAR-activated thrombin activity. With 4 human PARs identified so far, we wanted to determine the PAR expression profile in HMVEC by performing RT-PCR analysis. RT-PCR results showed that HMVEC expressed thrombin-sensitive PAR-1 and PAR-4, and thrombin-insensitive and trypsin-sensitive PAR-2. Under our assay conditions, RT-PCR could not amplify the expected band of the third thrombin-sensitive receptor, PAR-3, suggesting that HMVEC do not express PAR-3 or express PAR-3 at very low levels.
[0182] RT-PCR - HMVEC were cultured and RNA was isolated (TriReagent-BioMol) followed by DNase treatment and washing using the RNeasy Maxi kit (Qiagen). RT-PCR was performed using GC-rich PCR reagent (Invitrogen) and Superscript II reverse transcriptase (Invitrogen). Results were visualized by UV gel electrophoresis and imag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com