Low-temperature lipase and its gene sequence
A technology of lipase and low temperature, applied in genetic engineering, plant gene improvement, hydrolytic enzymes, etc., can solve the problem of low activation energy of low temperature lipase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Extraction of Pseudomonas marine total DNA
[0030] 1. Using Pseudomonas sp isolated from the sea, get 20 g of its fresh wet thalli and suspend in 10 ml of 50 mM Tris buffer (pH 8.0).
[0031] 2. Add a small amount of lysozyme and 8ml 0.25mM EDTA (pH8.0), mix well and place at 37°C for 20min.
[0032] 3. Add 2ml of 10% SDS and place at 55°C for 5min.
[0033] 4. Extract once each with equal volumes of phenol and chloroform.
[0034] 5. Take the last supernatant, add 2 times the volume of ethanol, and recover the DNA.
[0035] 6. Wash with 70% ethanol and absolute ethanol respectively.
[0036] 7. Dissolve the precipitate in 0.5ml TE buffer (pH 8.0, 10mM Tris, 1mM EDTA), add 3μl of 10mg / ml RNase, and incubate at 37°C for 1h.
[0037] 8. Extract once with equal volumes of phenol and chloroform respectively, and add 2 times the volume of ethanol to the supernatant to recover DNA.
[0038] 9. Wash the precipitate with 70% ethanol and absolute ethanol respecti...
Embodiment 2
[0040] Cloning of embodiment 2 low temperature lipase gene
[0041] 1. Take 0.5 μl of the total DNA solution of Pseudomonas marine as a template, and use the following oligonucleotide sequences as primers (including restriction sites of BamHI and HindIII), amplify the desired DNA fragments by conventional PCR techniques.
[0042] The primer sequences are as follows:
[0043] Primer 1: 5'-CTAAGCTTGTATCATTTTCGCGTCGT-3'
[0044] Primer 2: 5'-AGGGATCCGATGACTGATACATCGGC-3'
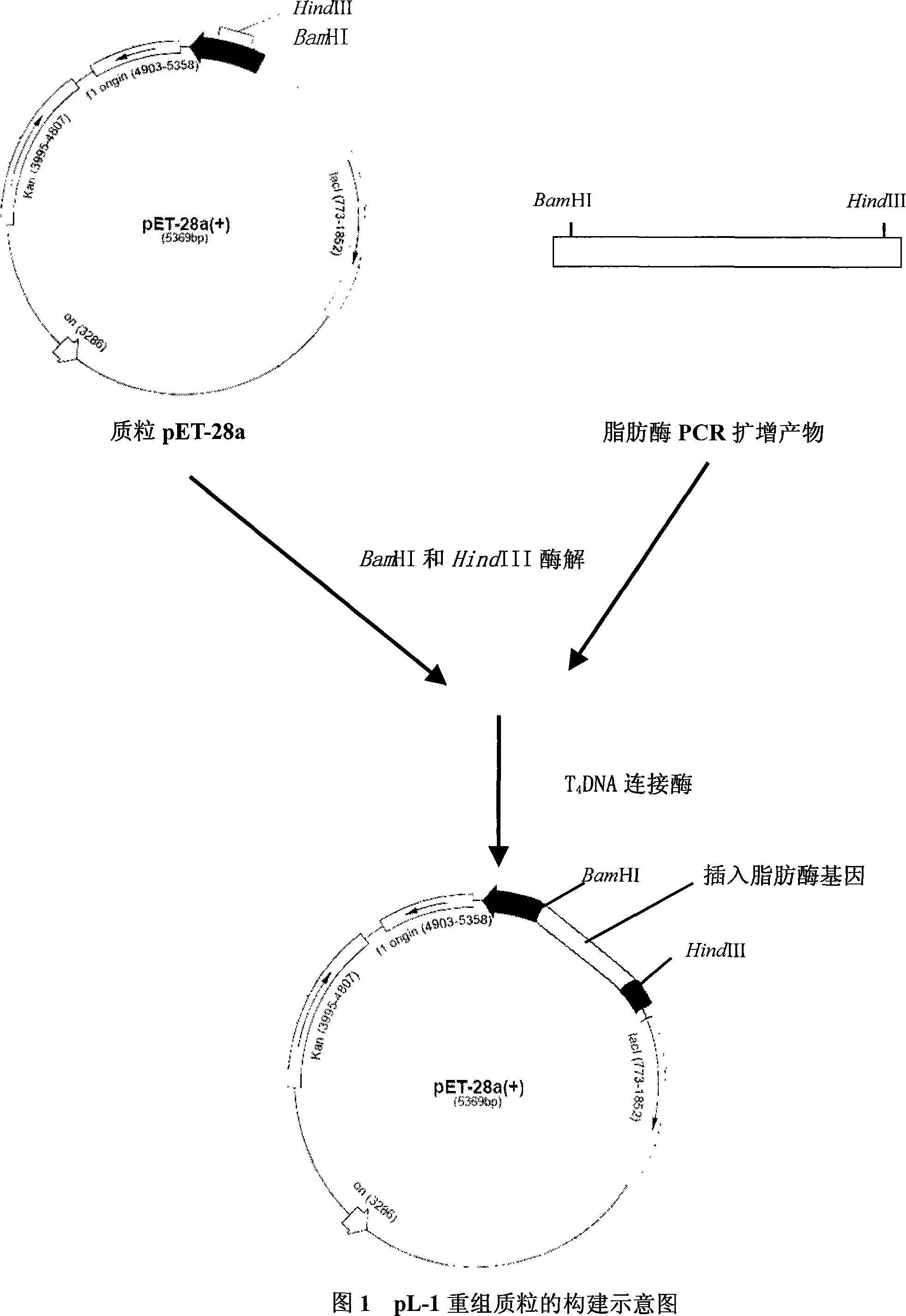
[0045] 2. Recover the PCR product, perform double digestion with restriction endonucleases BamHI and HindIII, and connect with the DNA of the plasmid pET-28a that has undergone the same digestion under the action of T4 DNA ligase to obtain the recombinant plasmid pL-1.
[0046] 4. Transform competent Escherichia coli BL21 with the recombinant plasmid. Spread on LB solid medium containing 50 μg / ml Kan (kanamycin) and olive oil. Incubate at 37°C for 12 hours, and then incubate at 60°C for 1-5 hours. If there ...
Embodiment 3
[0049] Example 3 High-efficiency expression of low-temperature lipase gene in engineering bacteria BL21LIP
[0050] The engineered strain BL21LIP was cultured in LB liquid medium containing kanamycin at 37°C for 16 hours with shaking, and then transferred to a fermenter. When the OD value of the bacteria reached 0.6-1.0, the expression was induced by adding ITPG with a final concentration of 0.6mM.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com