Oligonucleotide for targeted activation of chronic granulocyte leukaemia protein kinase PKR and application thereof
A chronic myeloid and oligonucleotide technology, applied in the field of nucleotide sequences for the treatment of chronic myeloid leukemia, to achieve the effect of easy tracking and observation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
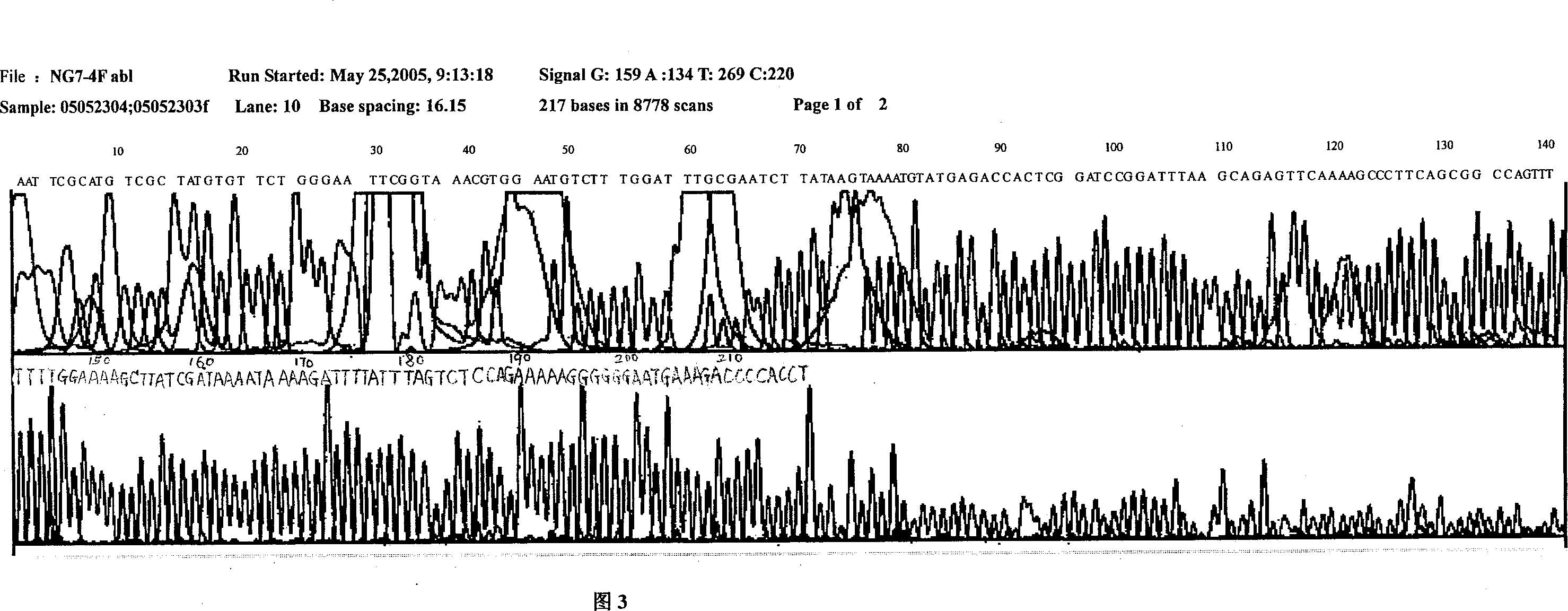
[0087] Example 1 Construction and Identification of pH1-BCR-ABL40as Recombinant Plasmid
[0088] 1.1 Obtaining of pH1-BCR-ABL40as insert fragment
[0089] Design SEQ ID NO 1 and SEQ ID NO 2 containing restriction endonuclease BamHI and HindIII sites at both ends respectively. Dissolve each fragment of 1OD in 50μL annealing buffer (10mM Tris, pH7.5~pH8.0, 50mM NaCl, 1mM EDTA), and mix well by inverting, and take 10μL each of SEQ ID NO 1 and SEQ ID NO 2 to 40as tube, placed in boiling water, naturally cooled and annealed into double strands, placed at 4°C for ligation reaction.
[0090] 1.2. Digestion and recovery
[0091] pSilencer3.1-H1
Hind III
10×K buffer
wxya 2 o
Totol
40μL
3μL
3μL
6μL
8μL
20 μL
[0092] The reactants were mixed and centrifuged and placed in a 37°C water bath overnight. After the enzyme digestion was completed, all the samples were subjected to 1% agarose gel electrophoresis, and the...
Embodiment 2
[0131] Example 2 Construction and identification of pMSCVeGFP retroviral vector
[0132] 2.1 PCR amplification of IRES2-EGFP vector eGFP fragment with primers Ec-GFP and Xh-GFP (Table 7)
[0133] ① PCR reaction system
[0134] 10×Buffer 3μL
[0135] 2mM dNTP 3μL
[0136] 25mM MgCl 2 1.8μL
[0137] Ec-GFP (10pmol / μL) 3μL
[0138] Xh-GFP (10pmol / μL) 3μL
[0139] IRES2-EGFP 0.2 μL
[0140] pfu Taq (5u / μL) 0.2μL
[0141] Add ddH 2 O up to 30μL
[0142] All the reagents were added into a 100 μL PCR tube, vortexed, centrifuged at 12,000 rpm for 30 sec, and put into a PCR machine for PCR reaction.
[0143] ② PCR thermal cycle reaction conditions
[0144] 94℃ 5min 1cycle
[0145] 94°C 1min
[0146] 55℃ 50sec 30cycles
[0147] 72℃ 40sec
[0148] 72℃ 5min 1cycle
[0149] After the thermal cycle is completed, centrifuge briefly at 12,000 rpm for 30 sec, take 5 μL of the reaction product, and perform electrophoresis analysis on 1.5% agarose gel.
[0150...
Embodiment 3
[0157] Example 3 Construction and identification of pMSCVeGFP-H1-BCR-ABL40AS recombinant retrovirus double expression vector
[0158] 3.1 Acquisition of insert fragments
[0159] The short fragment RNA expression cassette: H1-40as was amplified by PCR from the recombinant plasmid pH1-BCR-ABL40as, and the primers sa and Hd are shown in Table 7.
[0160] ① PCR reaction system
[0161] 10×Buffer 3μL
[0162] 2mM dNTP 3μL
[0163] 25mM MgCl 2 1.8μL
[0164] sa (10pmol / μL) 3μL
[0165] Hd (10pmol / μL) 3μL
[0166] 0.2 μL of each plasmid template
[0167] Taq (5u / μL) 0.2μL
[0168] ddH 2 0 to 30 μL
[0169] All the reagents were added into a 100 μL PCR tube, vortexed, centrifuged at 12,000 rpm for 30 sec, and put into a PCR machine for PCR reaction.
[0170] ② PCR thermal cycle reaction conditions
[0171] 94℃ 5min 1cycle
[0172] 94°C 1min
[0173] 55℃ 50sec 30cycles
[0174] 72℃ 40sec
[0175] 72℃ 5min 1cycle
[0176] After the thermal cycl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com