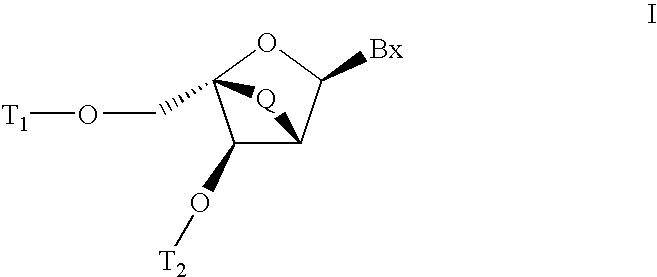
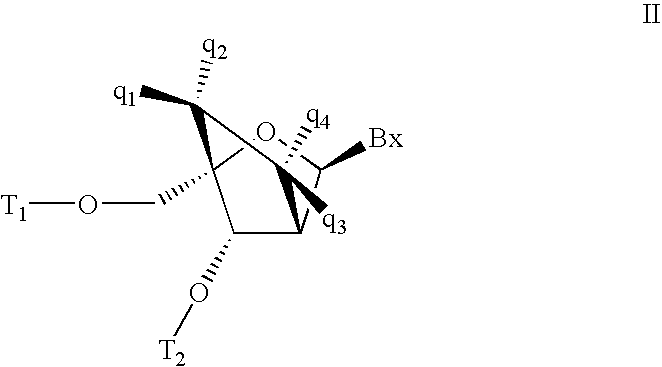
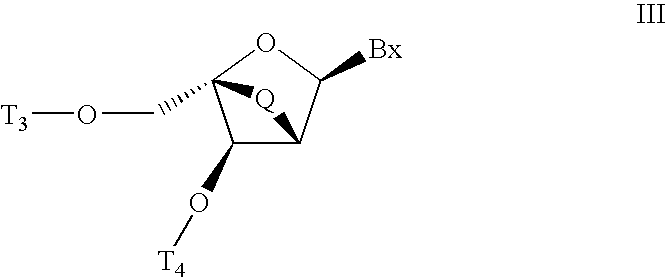
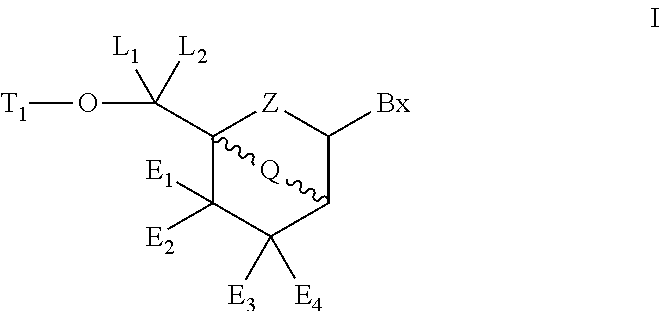
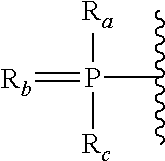
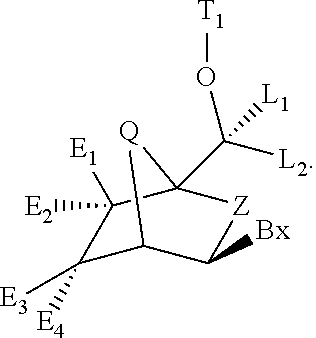
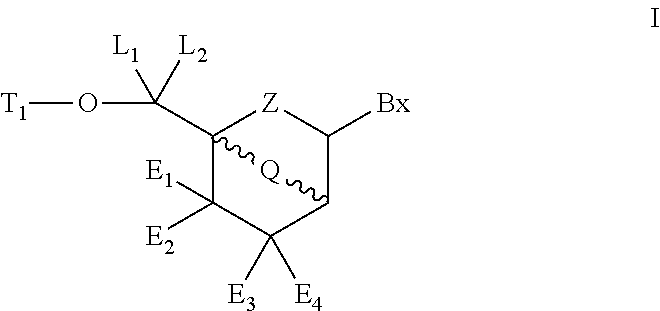
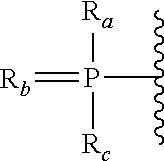
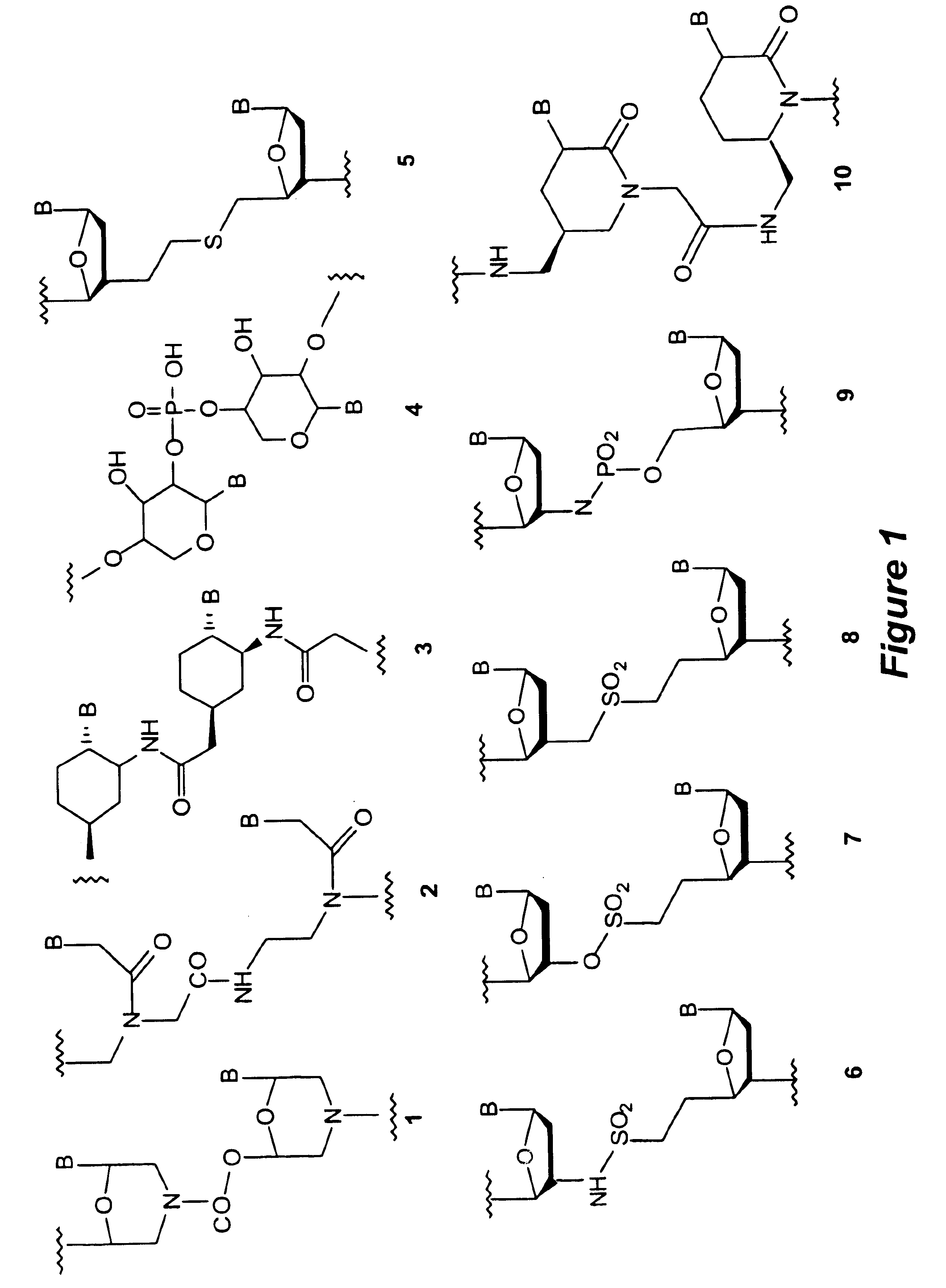
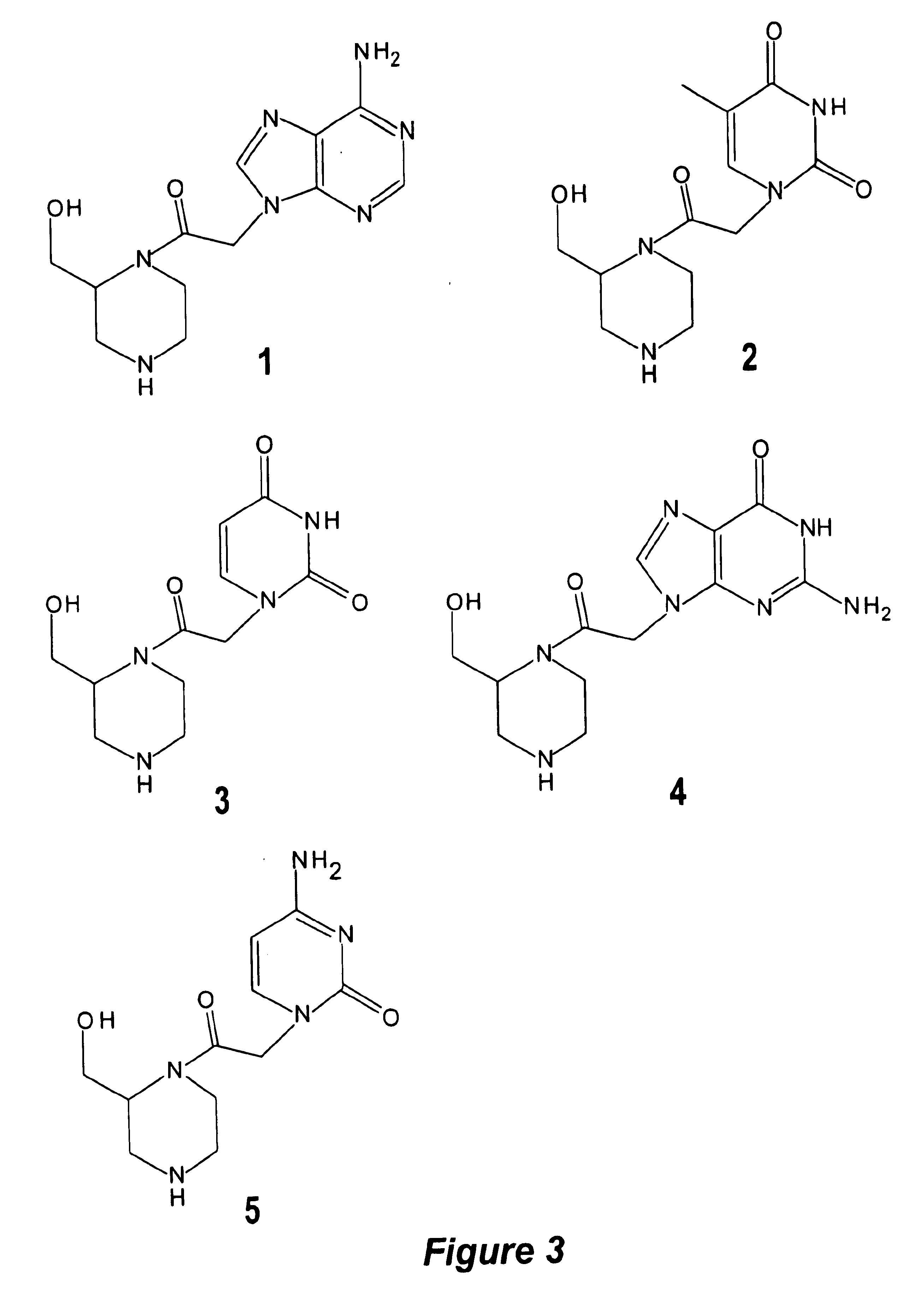
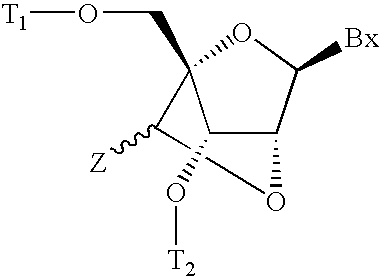
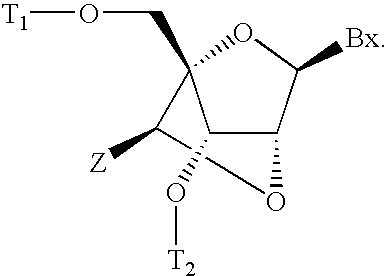
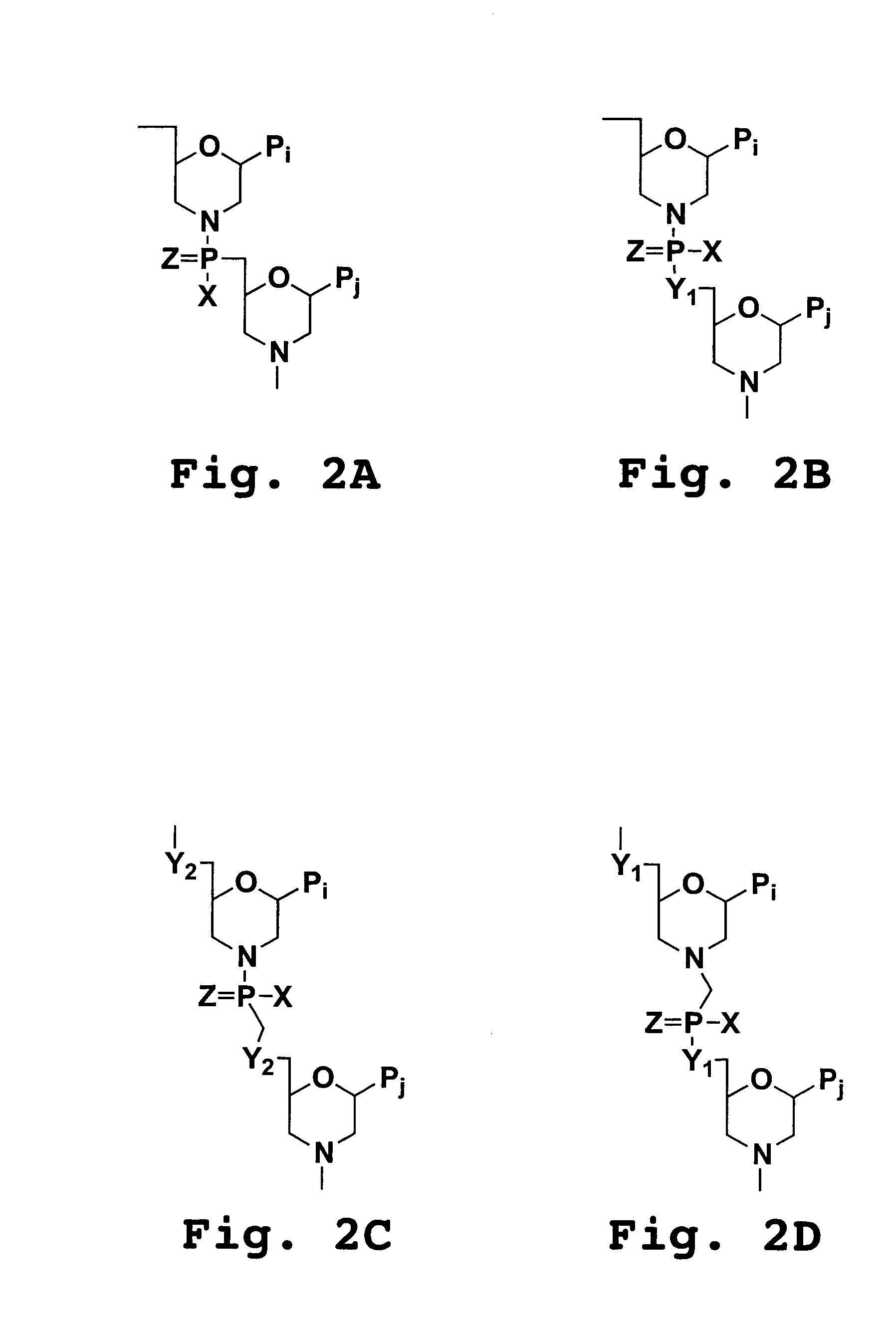Patents
Literature
Hiro is an intelligent assistant for R&D personnel, combined with Patent DNA, to facilitate innovative research.
47 results about "Nucleic acid analog" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
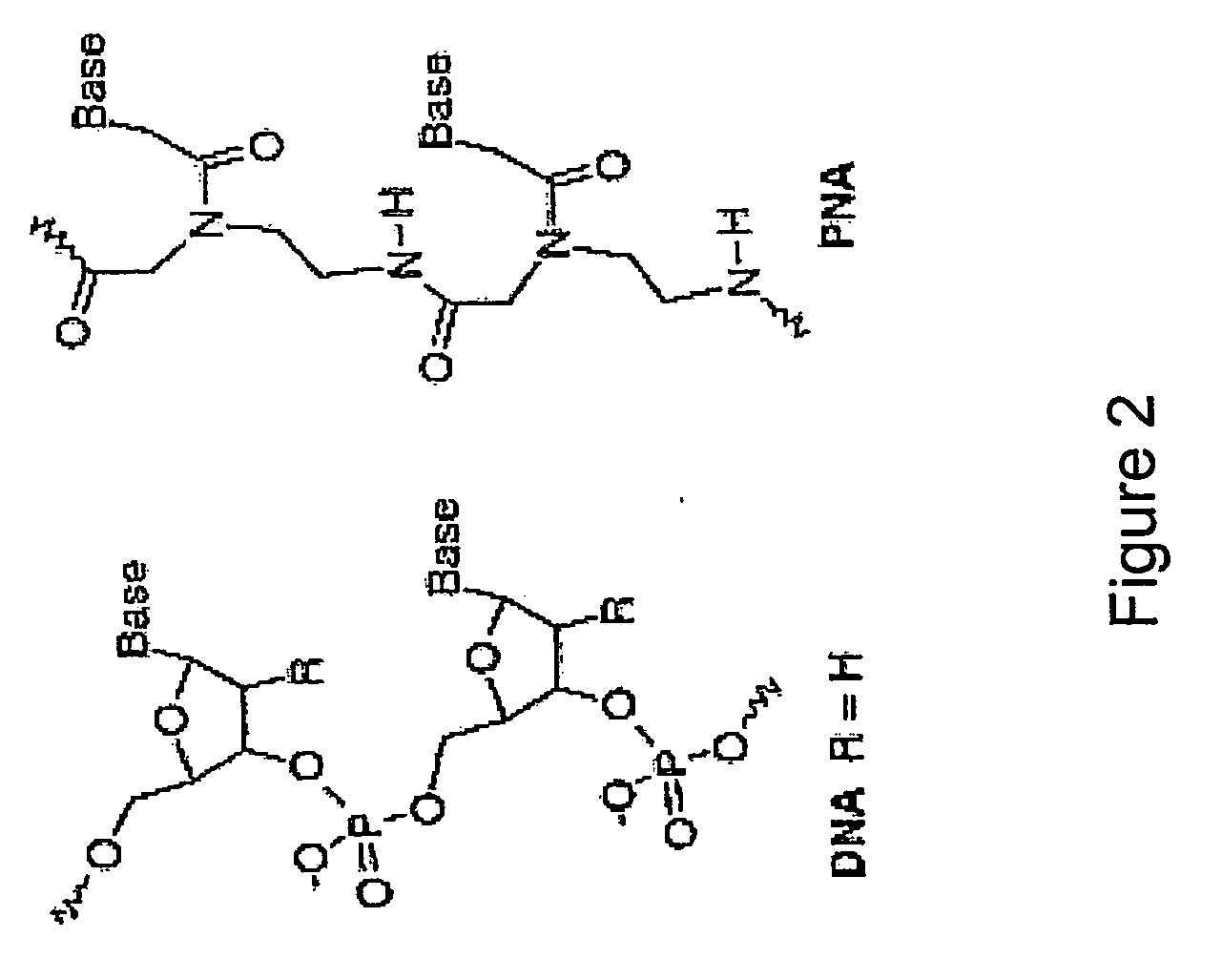
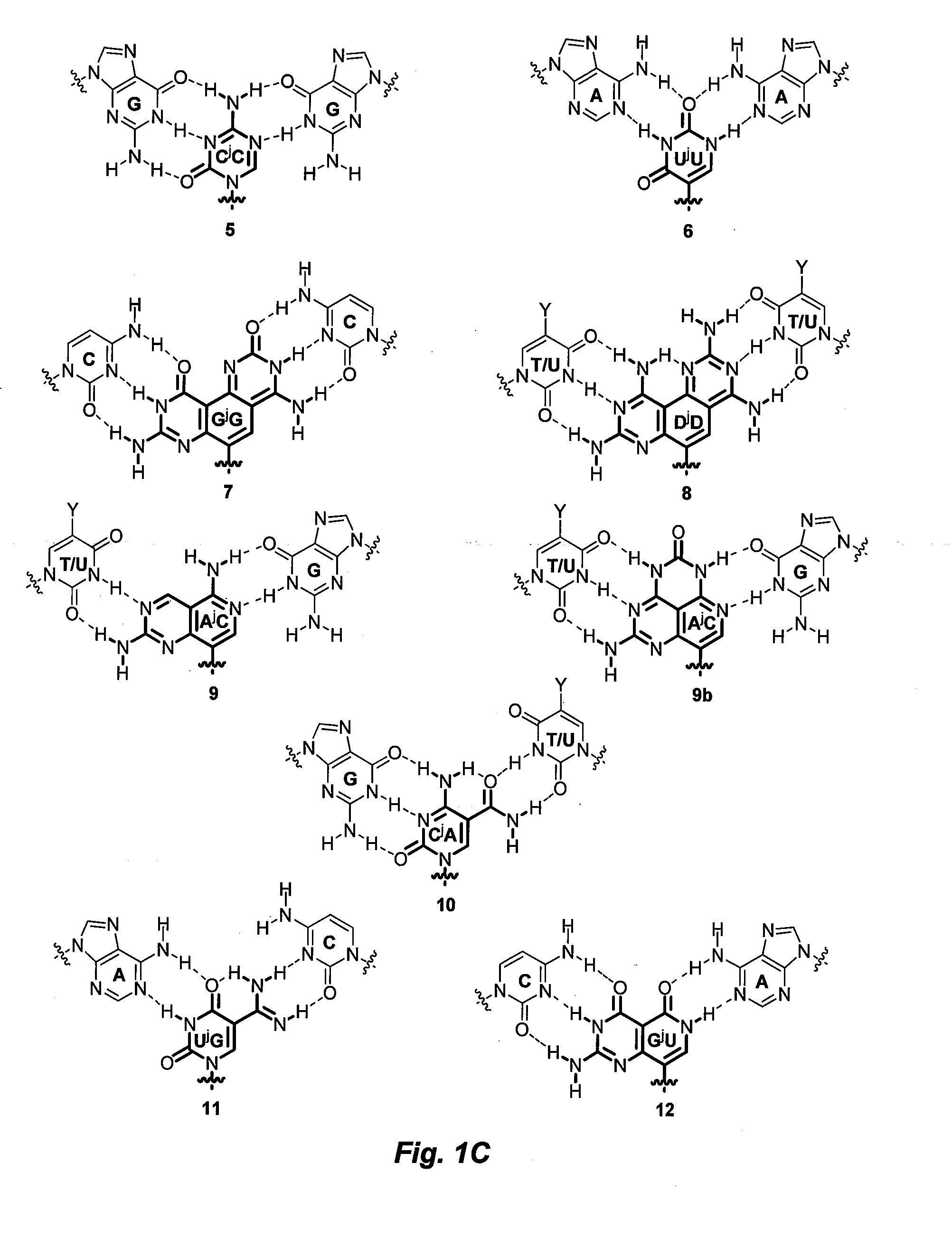
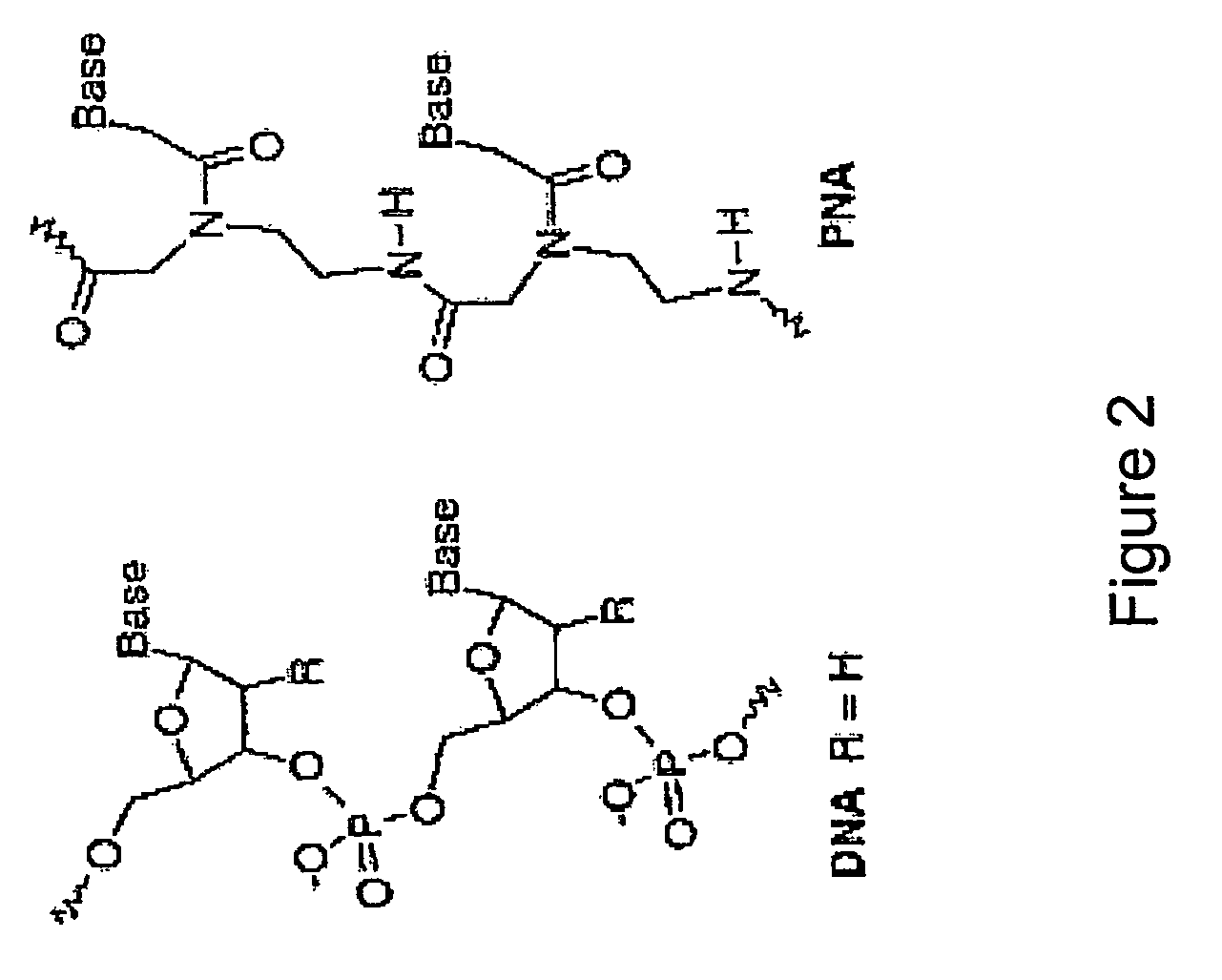
Nucleic acid analogue. Jump to navigation Jump to search This article needs attention from ... If this happens during DNA replication, a guanine will be inserted as the opposite base analog, and in the next DNA replication, that guanine will pair with a cytosine.
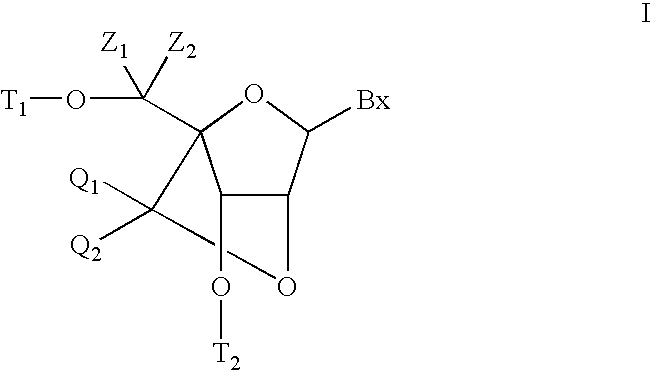
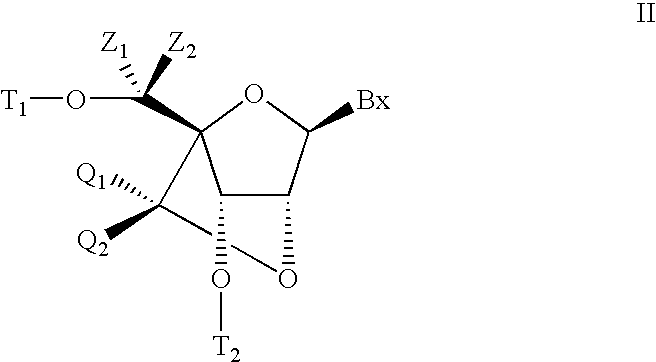
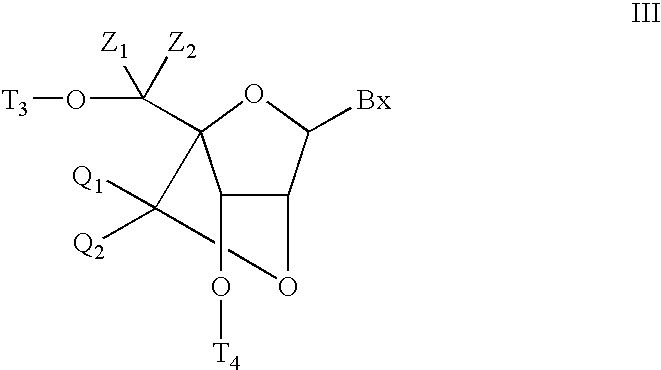
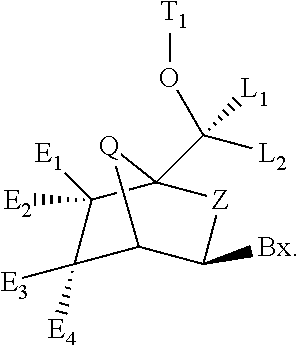
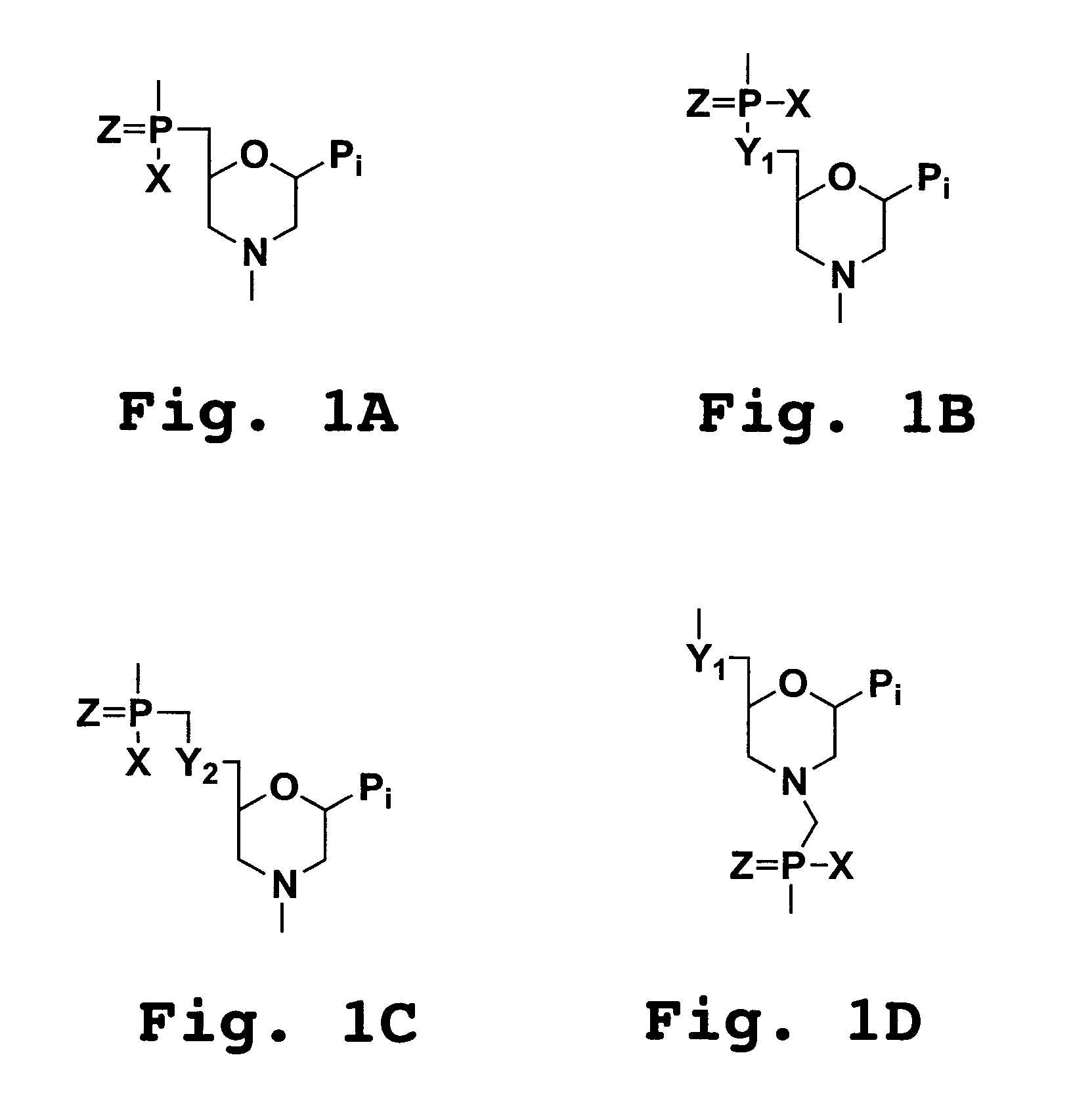
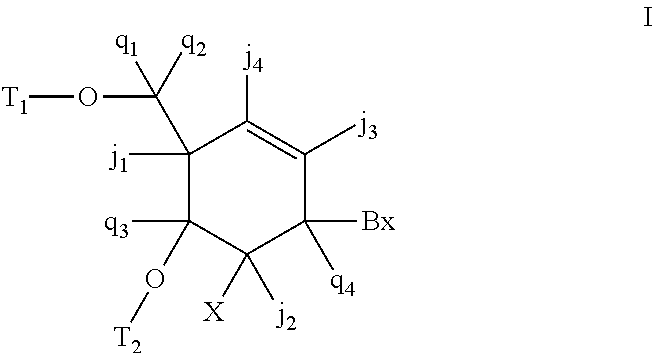
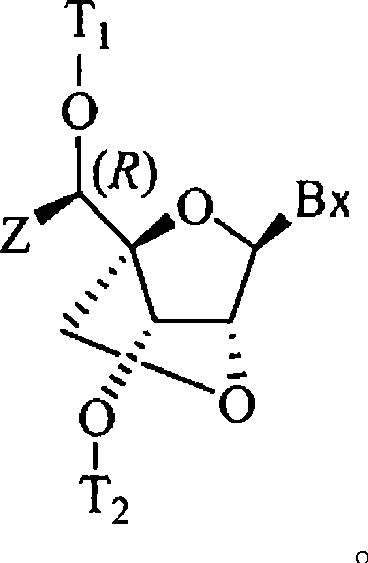
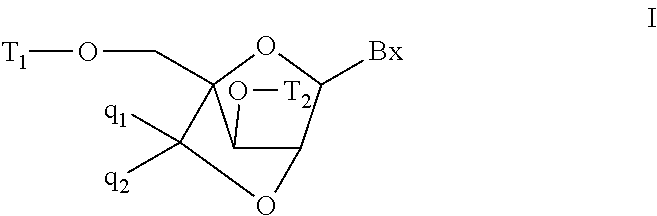
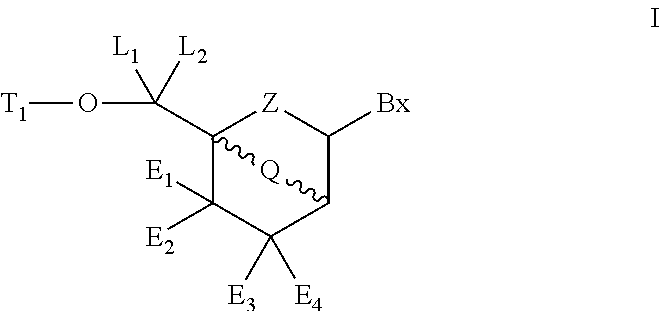
6-modified bicyclic nucleic acid analogs
The present invention provides 6-modified bicyclic nucleoside analogs and oligomeric compounds comprising these nucleoside analogs. In preferred embodiments the nucleoside analogs have either (R) or (S)-chirality at the 6-position. These bicyclicnucleoside analogs are useful for enhancing properties of oligomeric compounds including nuclease resistance.
Owner:IONIS PHARMA INC
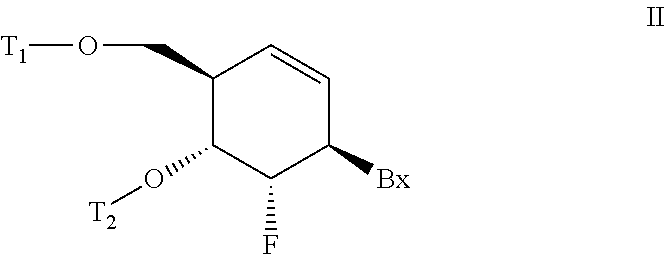
6-disubstituted or unsaturated bicyclic nucleic acid analogs
The present disclosure describes 6-disubstituted bicyclic nucleosides, oligomeric compounds prepared therefrom and methods of using the oligomeric compounds. More particularly, the 6-disubstituted bicyclic nucleosides each comprise a 2′-O—C(Ri)(R2)-4′ or 2′-O—C=(R3)(R.4)-4′ bridge wherein each R is, independently a substituent group and Ri and R2 include H. The 6-disubstituted bicyclic nucleosides are useful for enhancing properties of oligomeric compounds including nuclease resistance. In certain embodiments, the oligomeric compounds provided herein hybridize to a portion of a target RNA resulting in loss of normal function of the target RNA.
Owner:IONIS PHARMA INC
Carbocyclic bicyclic nucleic acid analogs
Provided herein are saturated and unsaturated carbocyclic bicyclic nucleosides, oligomeric compounds prepared therefrom and methods of using these oligomeric compounds. The saturated and unsaturated carbocyclic bicyclic nucleosides are useful for enhancing properties of oligomeric compounds including nuclease resistance.
Owner:IONIS PHARMA INC
Bis-modified bicyclic nucleic acid analogs
The present disclosure describes bis-modified bicyclic nucleosides and oligomeric compounds that can be prepared comprising at least one of these bis-modified bicyclic nucleosides. More particularly, the bis-modified bicyclic nucleosides have at least one substituent group at the 5′-methylene and on the bridge methylene and can be chiral. These bis-modified bicyclic nucleosides are expected to be useful for enhancing one or more property of oligomeric compounds including for example enhancing nuclease resistance.
Owner:IONIS PHARMA INC
Carbocyclic alpha-L-bicyclic nucleic acid analogs
The present invention provides novel carbocyclic α-L-bicyclic nucleosides and oligomeric compounds comprising at least one of these carbocyclic α-L-bicyclic nucleosides. The carbocyclic α-L-bicyclic nucleosides are useful for enhancing one or more properties of the oligomeric compounds they are incorporated into including nuclease resistance.
Owner:IONIS PHARMA INC
Bicyclic cyclohexose nucleic acid analogs
The present invention provides bicyclic cyclohexose nucleoside analogs and oligomeric compounds comprising these nucleoside analogs. These bicyclic nucleoside analogs are useful for enhancing properties of oligomeric compounds including nuclease resistance.
Owner:IONIS PHARMA INC
Bicyclic cyclohexose nucleic acid analogs
The present invention provides bicyclic cyclohexose nucleoside analogs and oligomeric compounds comprising these nucleoside analogs. These bicyclic nucleoside analogs are useful for enhancing properties of oligomeric compounds including nuclease resistance.
Owner:IONIS PHARMA INC
Compositions for enhancing transport of molecules into cells
ActiveUS7468418B2Easy to transportImprove bindingSpecial deliveryPeptide/protein ingredientsOligomerNucleic acid analog
Compositions and methods for enhancing delivery of molecules, e.g. biological agents, into cells are described. The composition is a conjugate of the biological agent, preferably a nucleic acid analog having a substantially uncharged backbone, covalently linked to a peptide transporter moiety as described. Conjugation of the peptide transporter to a substantially uncharged nucleic acid analog, such as a morpholino oligomer, is also shown to enhance binding of the oligomer to its target sequence and enhance antisense activity.
Owner:SAREPTA THERAPEUTICS INC
Multipartite high-affinity nucleic acid probes
InactiveUS6451588B1Accurately determineBioreactor/fermenter combinationsBiological substance pretreatmentsHybridization probeNucleic Acid Probes
The invention provides a collection of probes useful for hybridizing to a target nucleic acid. The probes associate with each other, binding with high affinity to the target nucleic acid, to form three-way junctions and other complexes. At least one of the probes in each collection includes a nucleic acid analog. Methods using the probes in hybridization and as primers are also provided.
Owner:APPL BIOSYSTEMS INC
Compositions for enhancing transport of molecules into cells
ActiveUS20090082547A1Improve bindingEasy to transportSpecial deliveryPeptidesOligomerNucleic acid analog
Compositions and methods for enhancing delivery of molecules, e.g. biological agents, into cells are described. The composition is a conjugate of the biological agent, preferably a nucleic acid analog having a substantially uncharged backbone, covalently linked to a peptide transporter moiety as described. Conjugation of the peptide transporter to a substantially uncharged nucleic acid analog, such as a morpholino oligomer, is also shown to enhance binding of the oligomer to its target sequence and enhance antisense activity.
Owner:SAREPTA THERAPEUTICS INC
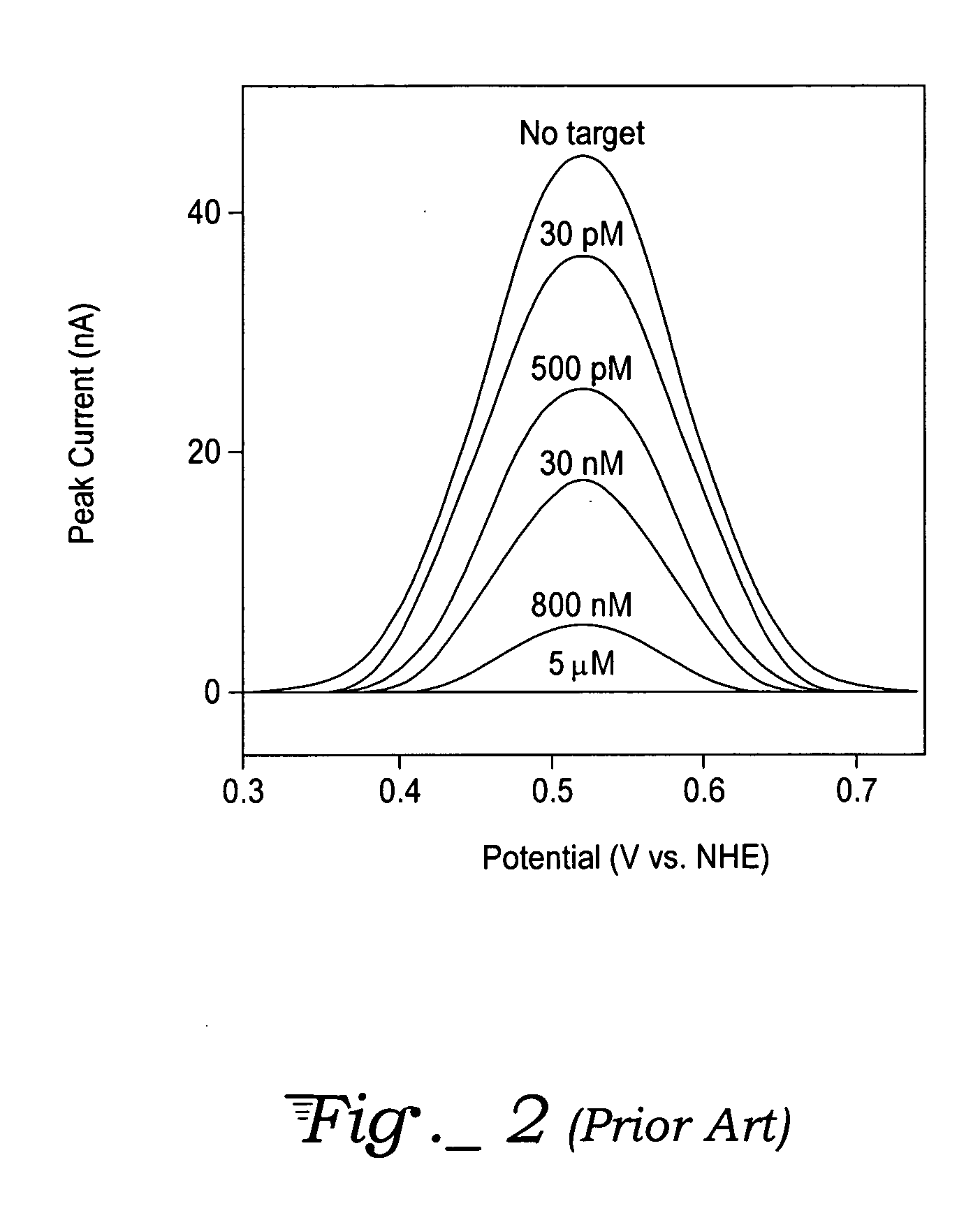
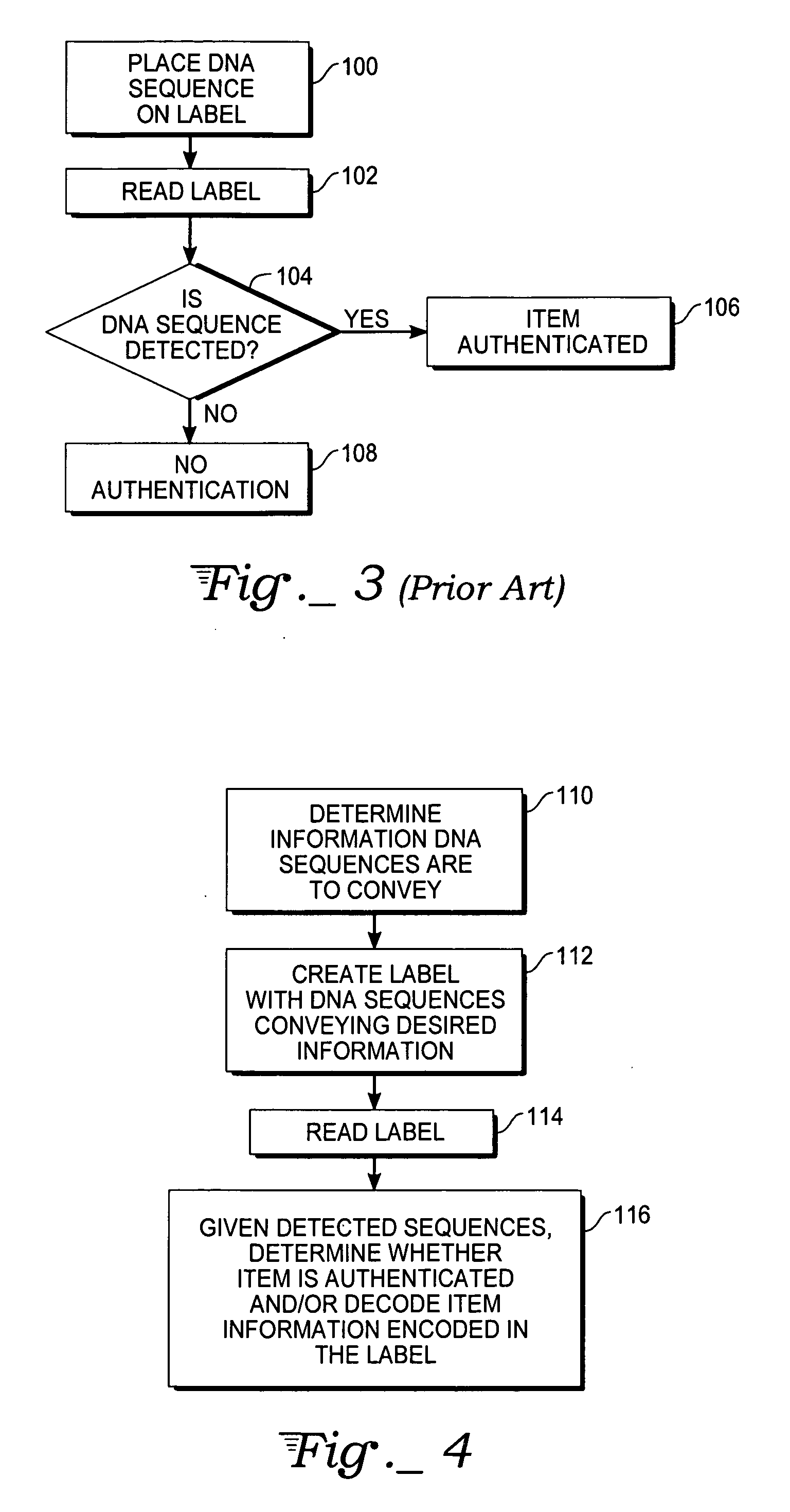
Method, apparatus, and system for authentication using labels containing nucleotide sequences
InactiveUS20060286569A1Microbiological testing/measurementBiological testingHash functionNucleotide sequencing
A method, label, and labeling system for labeling and authenticating an item are presented. At least one of a number of known nucleotide sequences associated with a predetermined amount of information is used as a label to be associated with an item. The label is then read with a reagentless sensor to detect the nucleotide sequence(s). The detected nucleotide sequence(s) is then associated with the appropriate information. The item is authenticated if the sensor detects the expected nucleotide sequence(s). The information in the DNA label may also be passed through a hash function or encrypted to further enhance security. The labels may also incorporate known non-natural nucleic acid analog sequences rather than nucleotide sequences, and a reader that reads known non-natural nucleic acid analog sequences may be employed.
Owner:BAR OR YUVAL +3
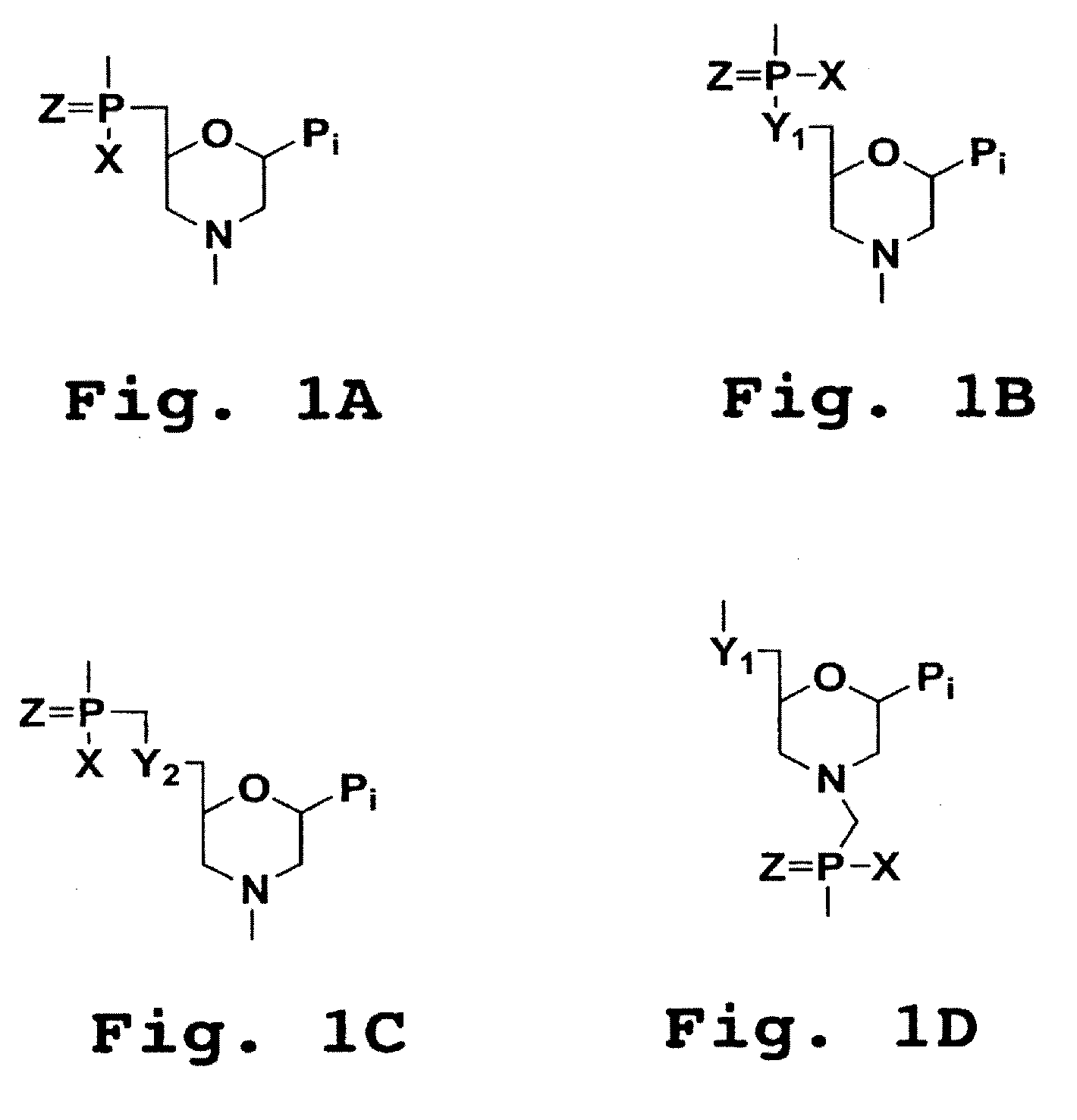
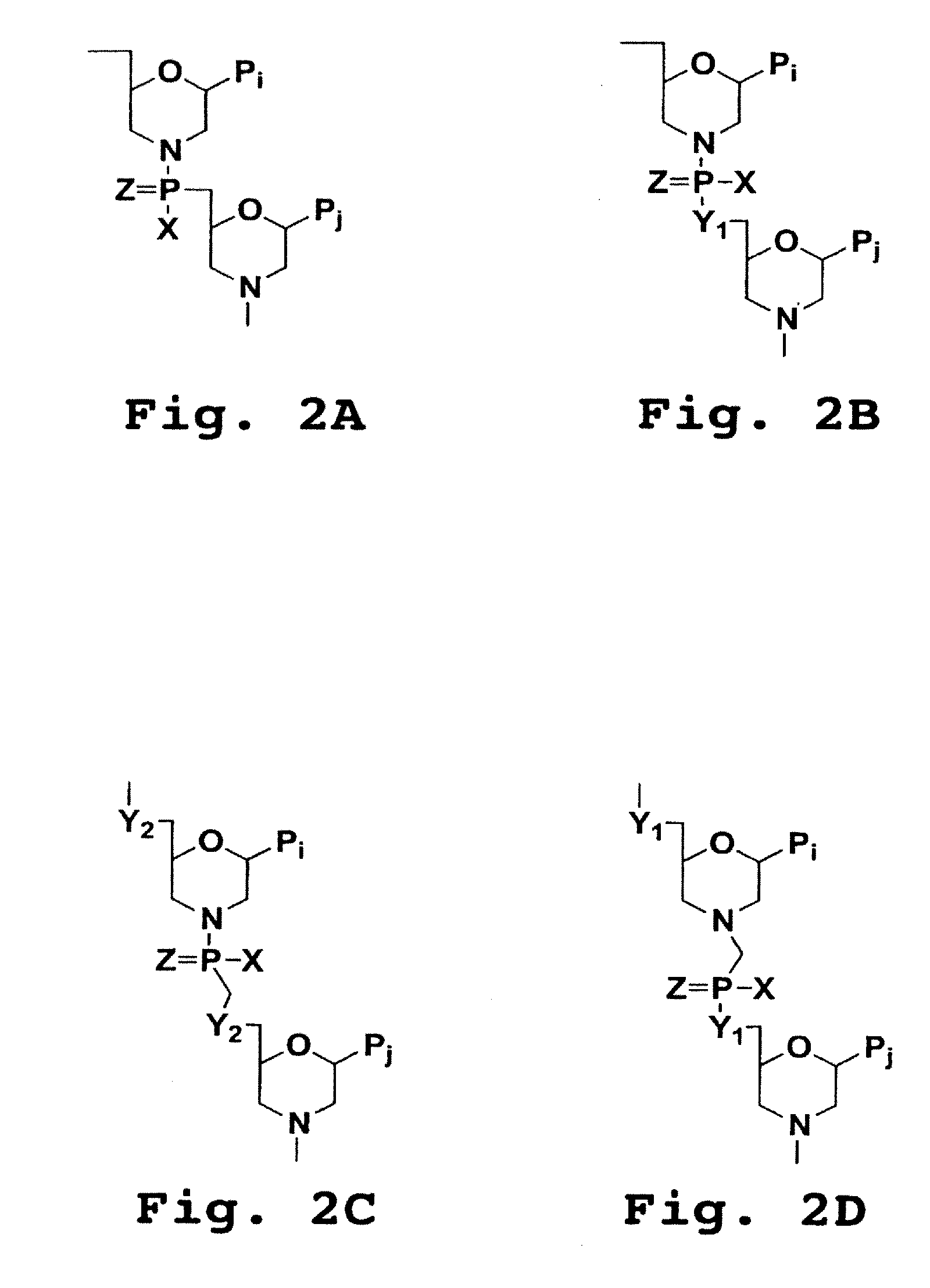
Piperazine-based nucleic acid analogs
InactiveUS6841675B1Quick assemblyEasy to assembleOrganic active ingredientsSugar derivativesOligomerNucleobase
A novel nucleoside analog is disclosed which comprises a piperazine ring in the place of the ring ribose or deoxyribose sugar. Monomers utilizing a broad variety of nucleobases are disclosed, as well as oligomers comprising the monomers disclosed herein linked by a variety of linkages, including amide, phosphonamide, and sulfonamide linkages. A method of synthesizing the nucleoside analogs is also disclosed.
Owner:LOS ALAMOS NATIONAL SECURITY
System for detecting polynucleotides
InactiveUS20060147958A1Increase rate of changeLow rate of changeMicrobiological testing/measurementFermentationOptical propertyPolynucleotide
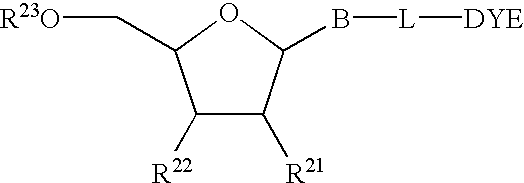
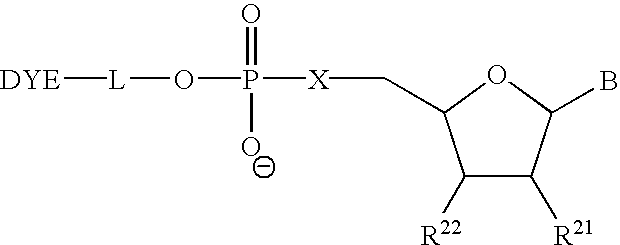
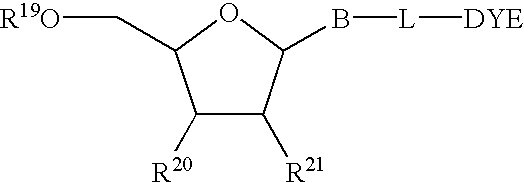
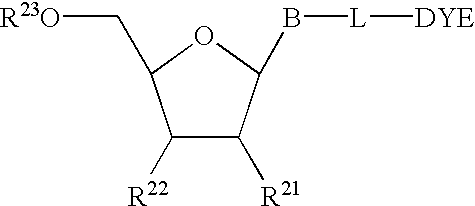
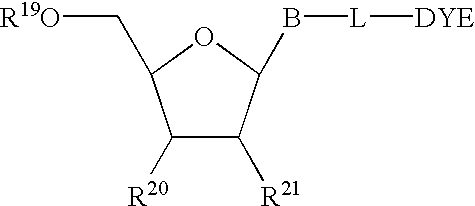
The present invention relates to methods for detecting the presence or amount of a target polynucleotide. A polynucleotide, target nucleic acid analog, and dye are combined to form a mixture. The optical property of the dye is observed after the mixture is exposed to a stimulating means. Optionally, after the stimulating means is employed, the mixture is compared to a reference value characteristic of the rate of change in the optical property of the dye in a similar mixture containing a known amount of a target polynucleotide / nucleic acid analog hybrid to determine a relative rate of change in the optical property. The change in a property of the mixture after exposure thereof to a stimulating means or the relative rate of change in the optical property of dye in the mixture is correlated with the presence or amount of the specified target polynucleotide in the sample.
Owner:INVESTIGEN
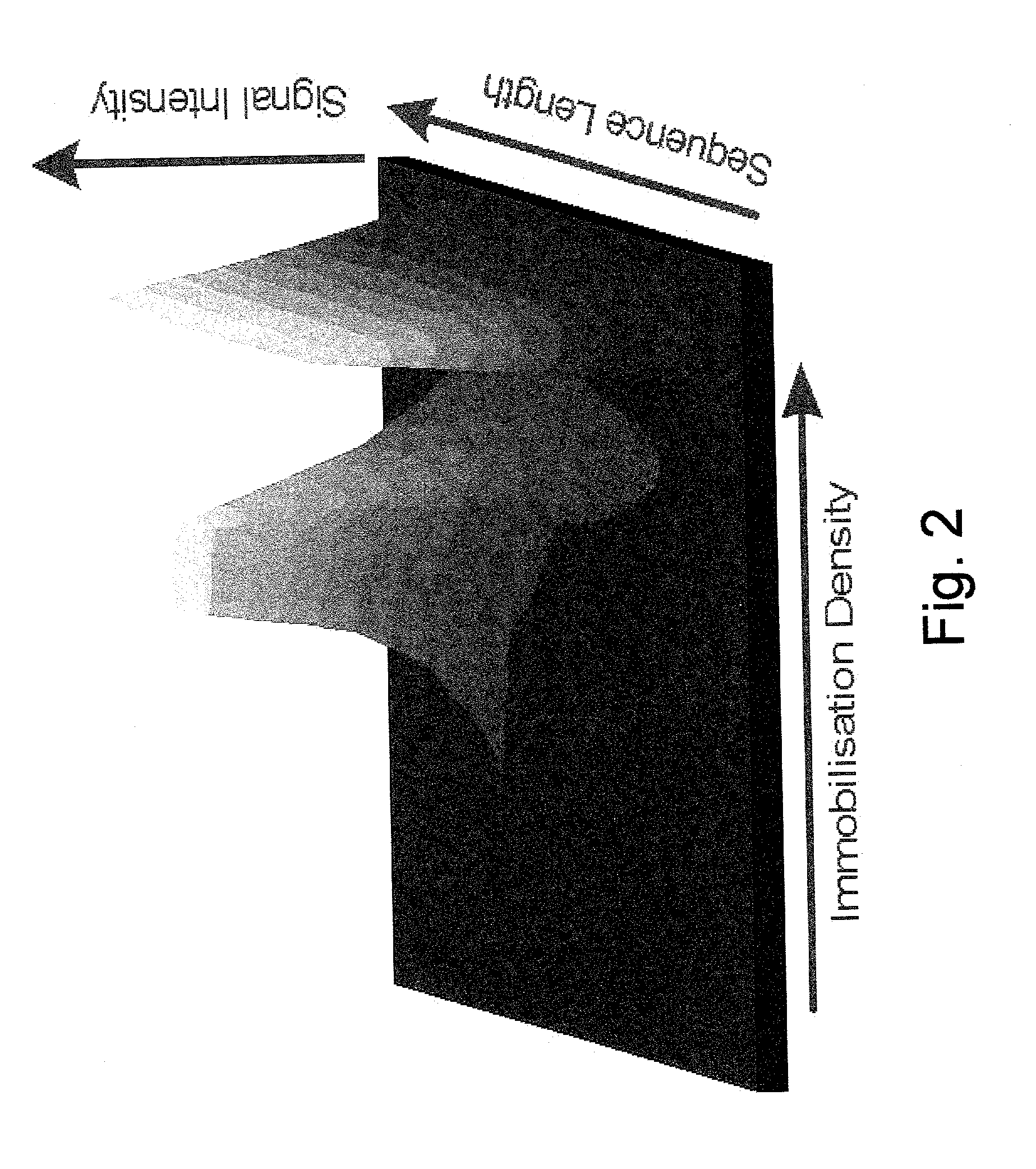
Gradient resolved information platform
InactiveUS20030055233A1Easy to identifyEasy to quantifyMaterial nanotechnologyBioreactor/fermenter combinationsNucleic Acid ProbesVolumetric Mass Density
The invention provides improved methods and devices for the detection and identification in a sample of one or more target molecules which bind to probe molecules, particularly to nucleic acid probe molecules. The improved method is based on contacting the sample with a surface that is coated with one or more gradients of probe molecules, particlarly nucleic acid or nucleic acid analog probe molecules that serve to bind target molecules in the sample, particularly nucleic acids having sequences that are complementary or partially complementary to one or more probe molecules. A probe gradient generated on the surface is formed by the variation of a physical, structural or functional property of the probes on the surface. The gradient is generated, e.g., by varying density of probe molecules bound to the surface, by varying probe sequence length, by varying probe sequence, by varying probe sequence type, by varying the orientational structure of probes, and by varying the concentration of label associated with probes. Determination of the location, speed and / or extent of hybridisation of a nucleic acid on such a gradient surface is useful to identify target molecules bound to probes and / or to quantitatively measure the amount of the target in a sample. Hybridisation of target molecules to a gradient of nucleic acid probe can be examined as a function of time and / or hybridisation conditions (e.g., temperature, salt concentration, etc.) The methods and devices of this invention employ gradient surfaces to bind to one or more target molecules, particularly nucleic acids (or target sequences) in a sample, detecting their presence in the sample and quantitating the amount of one or more of such targets in a sample.
Owner:KRULL ULRICH J
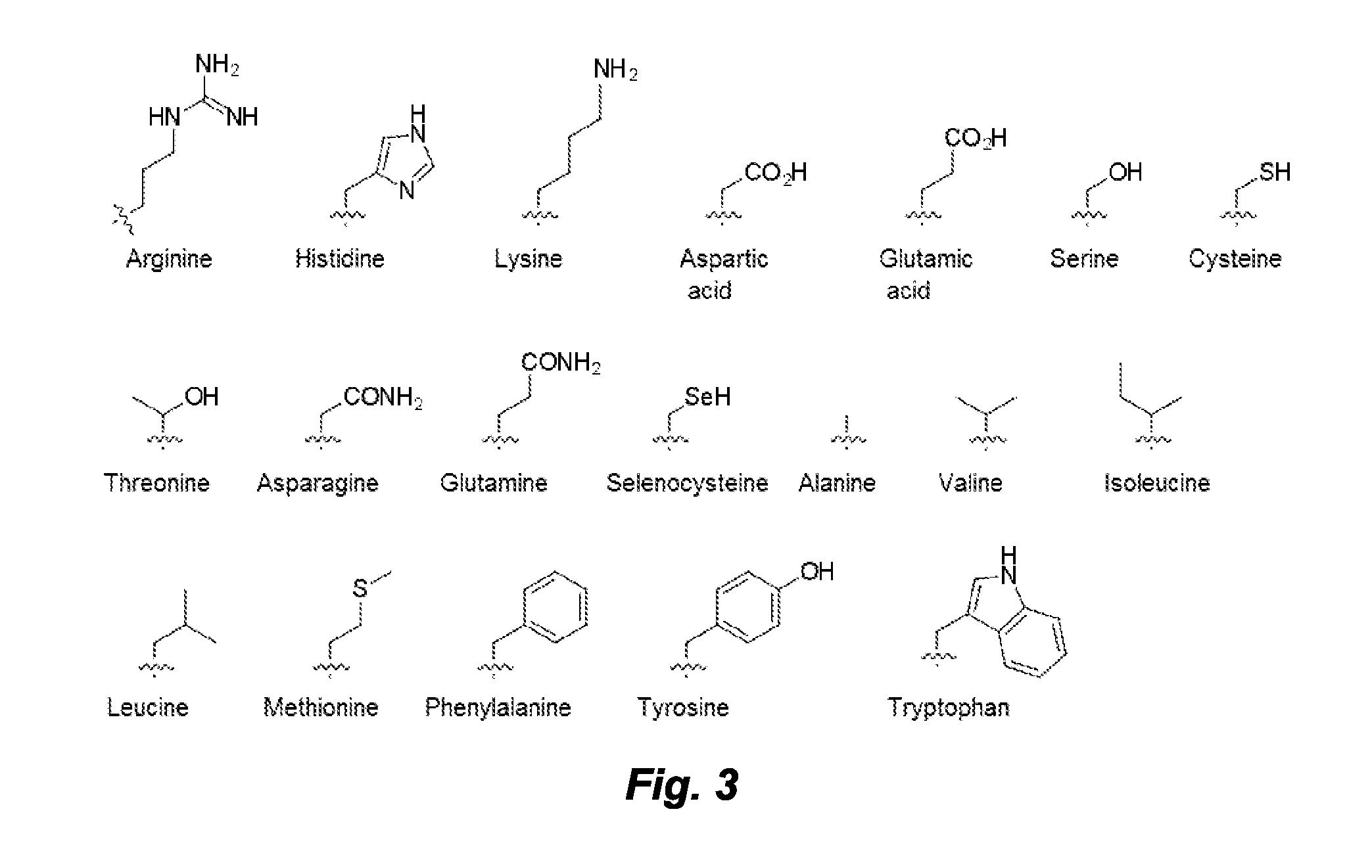
Divalent nucleobase compounds and uses therefor
Described herein are novel divalent nucleobases that each bind two nucleic acid strands, matched or mismatched when incorporated into a nucleic acid or nucleic acid analog backbone (a genetic recognition reagent, or genetic recognition reagent). In one embodiment, the genetic recognition reagent is a peptide nucleic acid (PNA) or gamma PNA (?PNA) oligomer. Uses of the divalent nucleobases and monomers and genetic recognition reagents containing the divalent nucleobases also are provided.
Owner:CARNEGIE MELLON UNIV
Left-Handed Gamma-Peptide Nucleic Acids, Methods of Synthesis and Uses Therefor
ActiveUS20170058325A1Reducing carboxylic acid groupMicrobiological testing/measurementPeptidesNano structuringSynthesis methods
A method of making optically pure preparations of chiral γPNA (gamma peptide nucleic acid) monomers is provided. Nano structures comprising chiral γPNA structures also are provided. Methods of amplifying and detecting specific nucleic acids, including in situ methods are provided as well as compositions and kits useful in those methods. Lastly, methods of converting nucleobase sequences from right-handed helical PNA, nucleic acid and nucleic acid analog structures to left-handed γPNA, and vice-versa, are provided.
Owner:CARNEGIE MELLON UNIV
Cyclohexenyl nucleic acids analogs
Owner:IONIS PHARMA INC
Enzyme-free isothermal exponential amplification of nucleic acids and nucleic acid analog signals
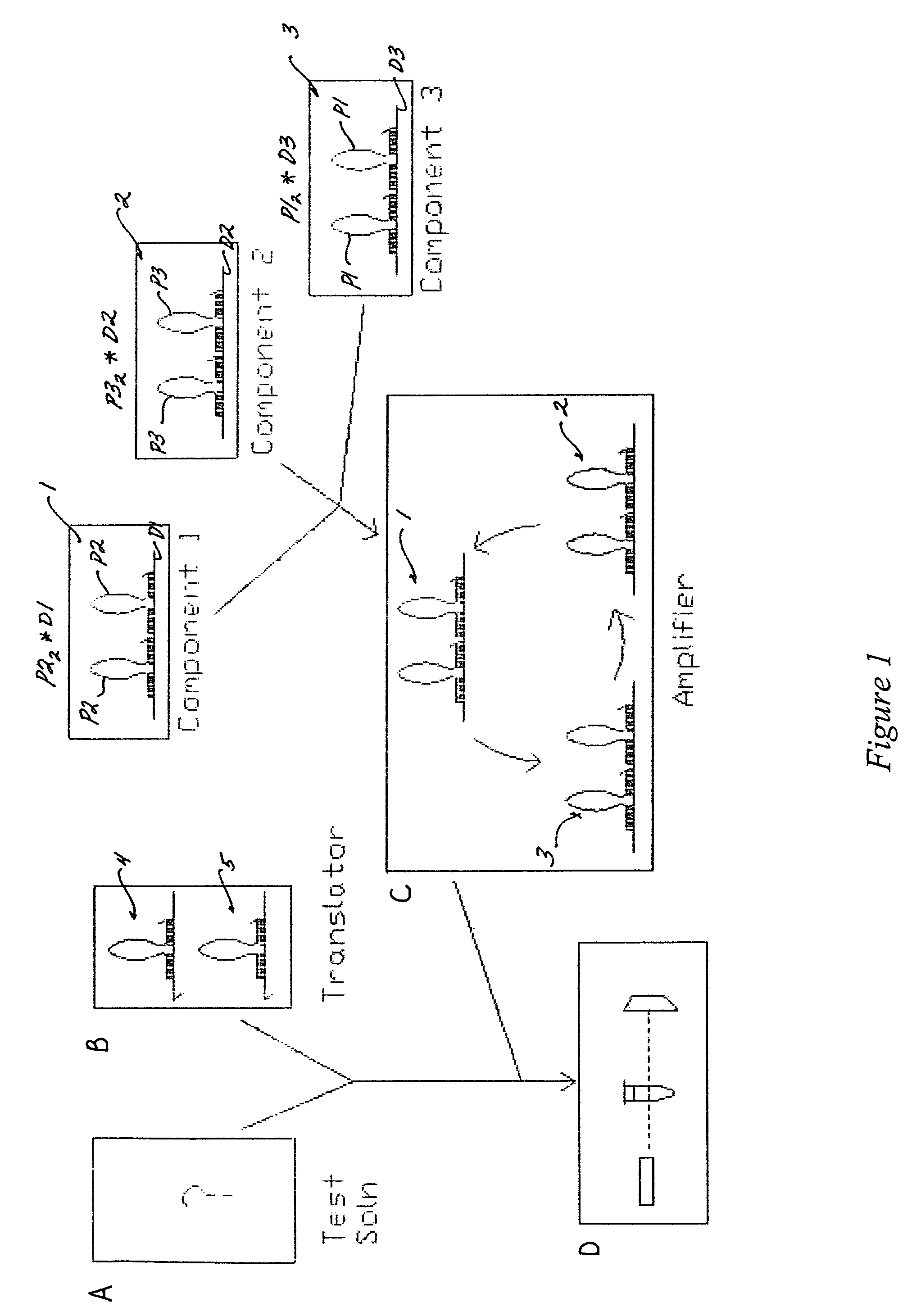
An enzyme-free, isothermal method of generating an amplification signal indicative of a target nucleic acid molecule is provided, as are compositions for performing such a method. An advantage of the detection system is that it is very sensitive, and can allow for the detection of a single target molecule in a sample.
Owner:CALIFORNIA INST OF TECH
Multipartite high-affinity nucleic acid probes
InactiveUS20030064402A1High affinitySugar derivativesMicrobiological testing/measurementHybridization probeNucleic Acid Probes
The invention provides a collection of probes useful for hybridizing to a target nucleic acid. The probes associate with each other, binding with high affinity to the target nucleic acid, to form three-way junctions and other complexes. At least one of the probes in each collection includes a nucleic acid analog. Methods using the probes in hybridization and as primers are also provided.
Owner:APPL BIOSYSTEMS INC
Blocking Agents Comprising Non-Natural Nucleic Acids and Detection Methods Using such Blocking Agents
InactiveUS20100075319A1Enhanced signalReducing and eliminating unwanted interactionMicrobiological testing/measurementHEXAPentanucleotide Repeats
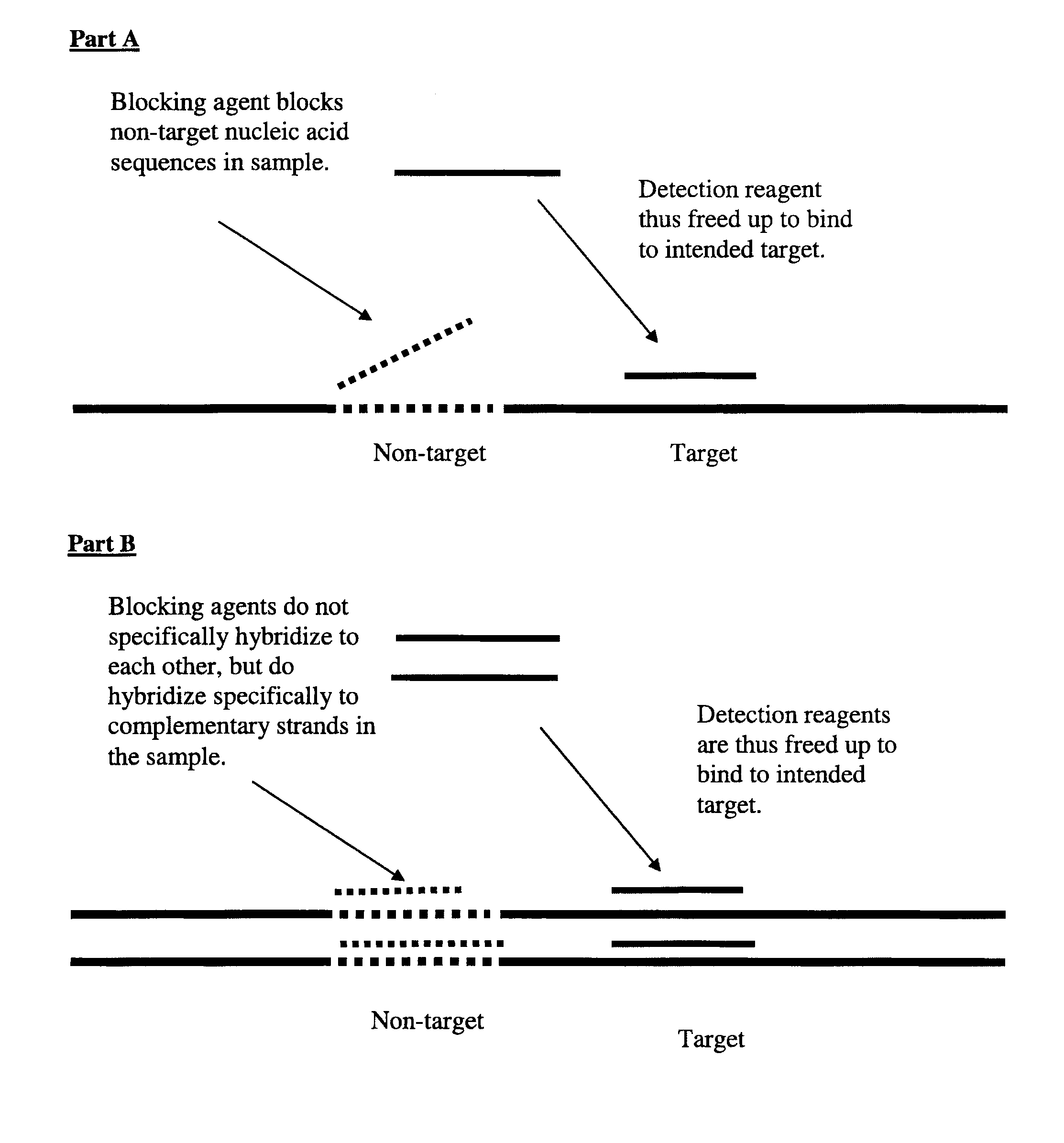
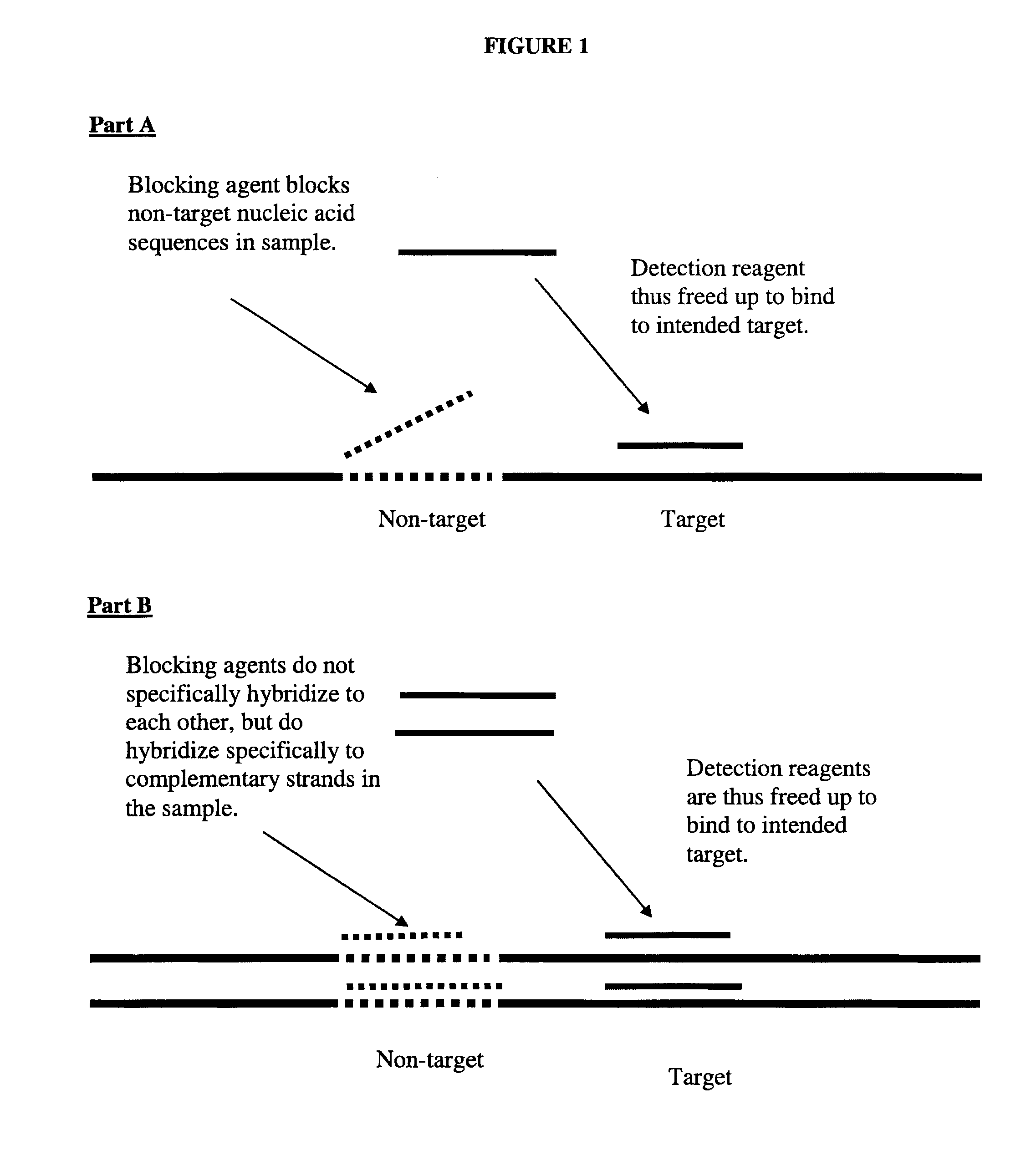
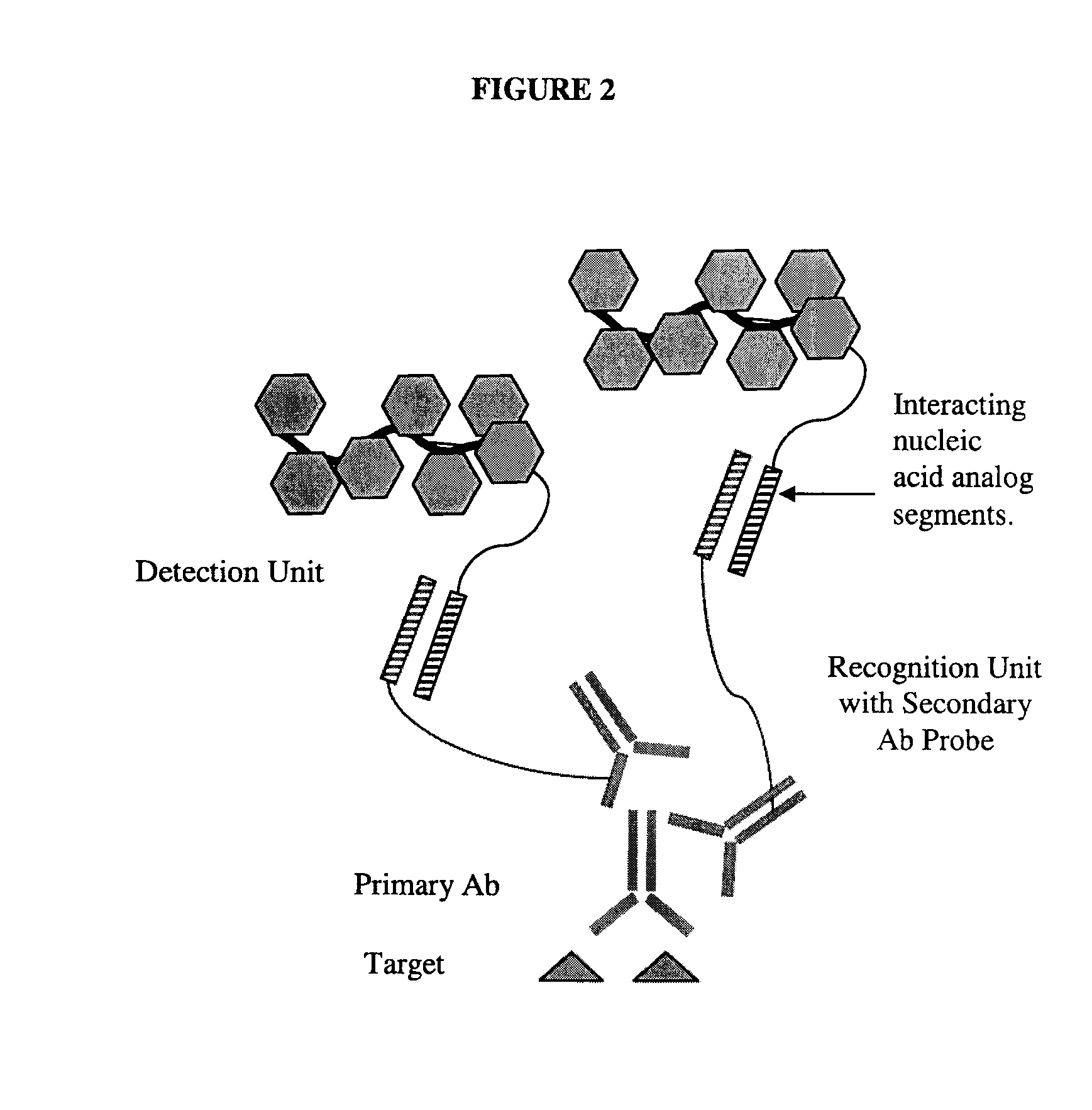
This invention relates to nucleic acid analog blocking agents that may reduce nonspecific interactions between components of a biological or chemical detection assay. The blocking agents may help to reduce the background observed in biological or chemical detection assays, such as in situ assays and blots, and may thus enhance the signal to noise in the assays. The invention also encompasses sets of nucleic acid analog segments, for instance, made from PNA and / or non-natural bases, which may act as blocking agents and / or detection reagents, reagent kits containing those sets, and related methods of detection. In some embodiments, the blocking agents are designed such that they block one or more sets of complementary strands of nucleic acids on a detection reagent or in a sample, but do not hybridize to each other. In some embodiments, the blocking agents may block genomic repeat sequences such as one or more of Alu repeats, Kpn repeats, di-nucleotide repeats, tri-nucleotide repeats, penta-nucleotide repeats, and hexa-nucleotide repeats.
Owner:DAKOAS
System for detecting polynucleotides
InactiveUS7745119B2Increase rate of changeLow rate of changeMicrobiological testing/measurementFermentationOptical propertyNucleotide
The present invention relates to methods for detecting the presence or amount of a target polynucleotide. A polynucleotide, target nucleic acid analog, and dye are combined to form a mixture. The optical property of the dye is observed after the mixture is exposed to a stimulating means. Optionally, after the stimulating means is employed, the mixture is compared to a reference value characteristic of the rate of change in the optical property of the dye in a similar mixture containing a known amount of a target polynucleotide / nucleic acid analog hybrid to determine a relative rate of change in the optical property. The change in a property of the mixture after exposure thereof to a stimulating means or the relative rate of change in the optical property of dye in the mixture is correlated with the presence or amount of the specified target polynucleotide in the sample.
Owner:INVESTIGEN INC
Methods for detecting chromosome aberrations
ActiveUS7306916B2Easy to useFast resultsSugar derivativesMicrobiological testing/measurementChromosome aberrationNucleic acid analog
The present invention relates to methods for detecting a change in chromosomal structure. These methods employ labeled probes that bind nucleic acids. For example, these probes may be comprised of nucleic acids or nucleic acid analogs and a detectable label.
Owner:AGILENT TECH INC
Divalent nucleobase compounds and uses therefor
Described herein are novel divalent nucleobases that each bind two nucleic acid strands, matched or mismatched when incorporated into a nucleic acid or nucleic acid analog backbone (a genetic recognition reagent, or genetic recognition reagent). In one embodiment, the genetic recognition reagent is a peptide nucleic acid (PNA) or gamma PNA (?PNA) oligomer. Uses of the divalent nucleobases and monomers and genetic recognition reagents containing the divalent nucleobases also are provided.
Owner:CARNEGIE MELLON UNIV
Enzyme-free isothermal exponential amplification of nucleic acids and nucleic acid analog signals
InactiveUS20050227259A1Increased secondary structureSugar derivativesMicrobiological testing/measurementNucleic acid analogEnzyme
An enzyme-free, isothermal method of generating an amplification signal indicative of a target nucleic acid molecule is provided, as are compositions for performing such a method. An advantage of the detection system is that it is very sensitive, and can allow for the detection of a single target molecule in a sample.
Owner:CALIFORNIA INST OF TECH
Bis-modified bicyclic nucleic acid analogs
ActiveUS20090156792A1Improve propertiesEsterified saccharide compoundsBiocideMethyl groupNucleic acid analog
Owner:IONIS PHARMA INC
5'-modified bicyclic nucleic acid analogs
The present invention provides 5'-modified bicyclic nucleoside analogs and oligomeric compounds comprising at least one of these nucleoside analogs. In preferred embodiments the nucleoside analogs have either (R) or (S)-chirality at the 5'-carbon. These bicyclic nucleoside analogs are useful for enhancing properties of oligomeric compounds including for example enhanced nuclease resistance.
Owner:IONIS PHARMA INC
6-Disubstituted Or Unsaturated Bicyclic Nucleic Acid Analogs
ActiveUS20110053881A1Suppress gene expressionBiocideSugar derivativesOrganic chemistryNucleic acid analog
The present disclosure describes 6-disubstituted bicyclic nucleosides, oligomeric compounds prepared therefrom and methods of using the oligomeric compounds. More particularly, the 6-disubstituted bicyclic nucleosides each comprise a 2′-O—C(Ri)(R2)-4′ or 2′-O—C=(R3)(R.4)-4′ bridge wherein each R is, independently a substituent group and Ri and R2 include H. The 6-disubstituted bicyclic nucleosides are useful for enhancing properties of oligomeric compounds including nuclease resistance. In certain embodiments, the oligomeric compounds provided herein hybridize to a portion of a target RNA resulting in loss of normal function of the target RNA.
Owner:IONIS PHARMA INC
Bicyclic cyclohexose nucleic acid analogs
The present invention provides bicyclic cyclohexose nucleoside analogs and oligomeric compounds comprising these nucleoside analogs. These bicyclic nucleoside analogs are useful for enhancing properties of oligomeric compounds including nuclease resistance.
Owner:IONIS PHARMA INC
6-modified bicyclic nucleic acid analogs
ActiveUS20070249049A1Suppress gene expressionSugar derivativesGenetic material ingredientsNucleaseChirality
The present invention provides 6-modified bicyclic nucleoside analogs and oligomeric compounds comprising these nucleoside analogs. In preferred embodiments the nucleoside analogs have either (R) or (S)-chirality at the 6-position. These bicyclicnucleoside analogs are useful for enhancing properties of oligomeric compounds including nuclease resistance.
Owner:IONIS PHARMA INC
Morphatides: novel shape and structure libraries
InactiveUS6838238B1Effectively “ evolve ”Simple structureSilicon organic compoundsSugar derivativesBiologyNucleic acid analog
This invention provides a method for identifying one or more complexes from a library of complexes, wherein said complex or complexes are selected for their ability to perform a preselected or desired function on a target molecule or by having a pre-selected structure, each complex being designated a morphatide, said method comprising: (a) preparing a library of morphatides, comprised of: (i) a scaffolding component selected from the group consisting of nucleic acid, nucleic acid like molecule or nucleic acid analog having one or more regions of randomized sequence; (ii) one or more linker components; and (iii) one or more agent molecules or type of agent molecules, linked to the scaffolding component by one or more type of linker components; and (b)screening the library of morphatides prepared in step (a) by contacting, binding, or associating the morphatides with one or more suitable target molecules upon which a morphatide performs a preselected or desired function or to which a morphatide binds or associates through a pre-selected structure of said morphatide under conditions permitting said morphatide to perform said preselected or desired function on said target molecules or permitting said morphatide to bind or associate with said target molecules through the preselected structure; (c) separating the morphatides performing the preselected or desired function or binding or associating through the preselected structure, from the library of morphatides and target molecules; thereby identifying one or more complexes from a library of complexes, wherein said complex or complexes are selected for their ability to perform a preselected or desired function on a target molecule or by having a pre-selected structure.
Owner:LIFE TECH CORP
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com