Methods for predicting drug responsiveness in cancer patients
a cancer patient and drug responsiveness technology, applied in the field of methods for predicting drug responsiveness in cancer patients, can solve the problems of cancer cells often developing resistance to a previously effective therapy, often losing critical time,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
ation of Biomarkers of Sensitivity and Resistance to Dovitinib
[0402]The drug dovitinib was submitted to NCI for testing in their NCI60 panel of cell lines. The measured growth inhibition, expressed as −log(GI50) values for cell lines in the panel, are as shown in Table 1:
TABLE 1Measured growth inhibition by dovitinib in NCI60 panel.Cell line-log(GI50)CCRF-CEM6.23HL-60(TB)5.97K-5626.26MOLT-46.48RPMI-82265.83SR6.35A549 / ATCC6.25EKVX5.12HOP-625.84HOP-926.45NCI-H2266.45NCI-H235.61NCI-H322M6.12NCI-H4606.33NCI-H5225.5COLO 2055.65HCC-29985.88HCT-1166.38HCT-156.37HT295.85KM127.38SW-6206.26SF-2685.66SF-2955.22SF-5396.37SNB-195.58SNB-756.01U2516.26LOX IMVI6.06MALME-3M6.31M146.41MDA-MB-4355.97SK-MEL-26.05SK-MEL-285.83SK-MEL-55.75UACC-2575.47UACC-625.44IGROV16.6OVCAR-35.17OVCAR-46.24OVCAR-55.75OVCAR-85.84NCI / ADR-RES5.57SK-OV-36.29786-06.14A4986.87ACHN6.48CAKI-16.33RXF 3936.66SN12C6.17TK-105.11UO-316.66PC-35.95DU-1455.94MCF75.93MDA-MB-231 / ATCC5.63HS 578T6.01BT-5495.98T-47D5MDA-MB-4685.59
[0403]Bas...
example 2
Versus Sorafenib for Third-Line Targeted Treatment of Patients with Metastatic Renal Cell Carcinoma: An Open-Label, Randomized Phase 3 Trial (ClinicalTrials.Gov, Number NCT01223027)
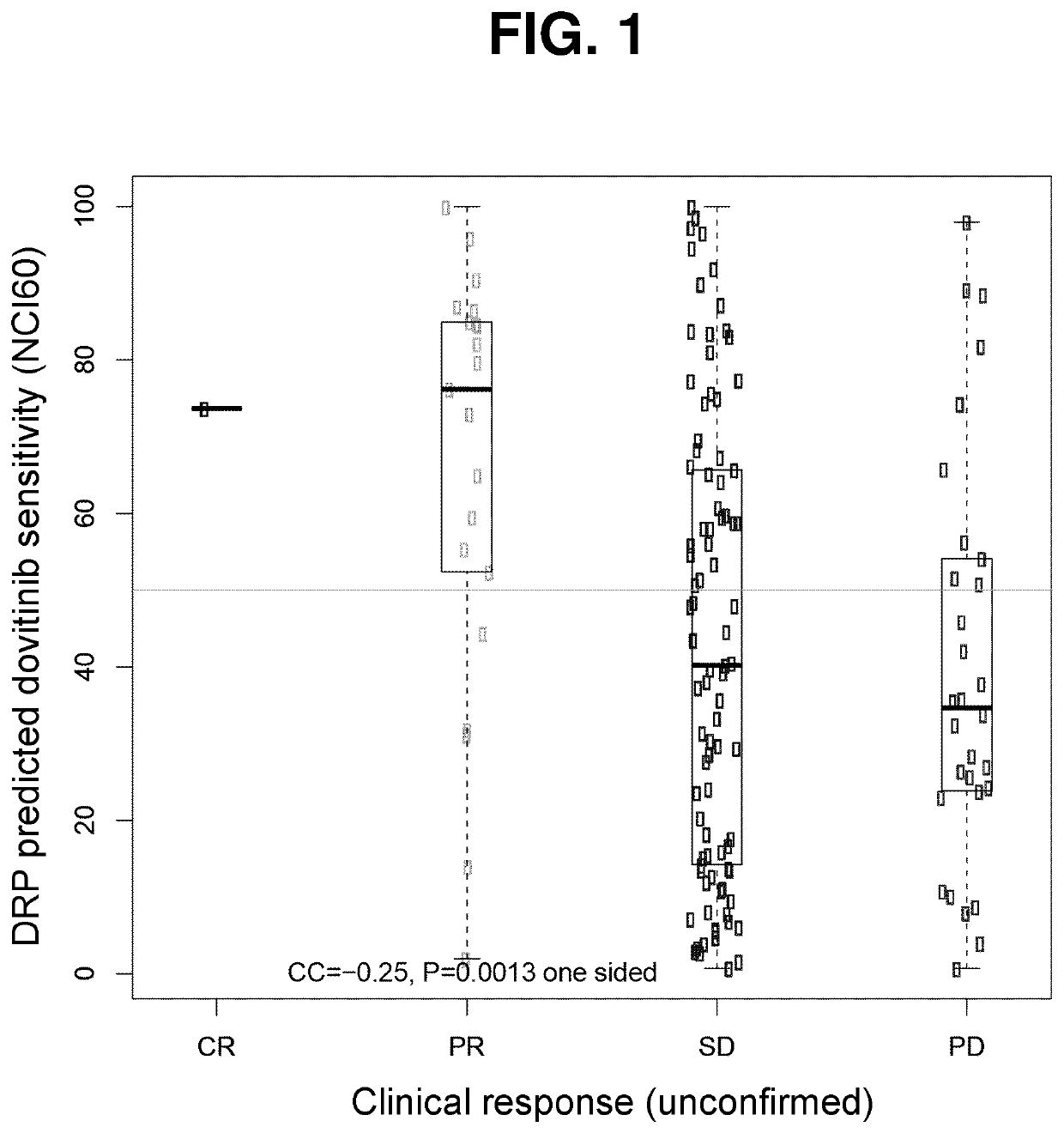
[0404]Gene expression data (Affymetrix) and clinical response data was obtained from 73 patients in the dovitinib arm. The DRP was applied to calculate a score for each patient (mean of genes in Table 2 minus mean of genes in Table 3). The score for each patient was compared to a reference population with the same diagnosis. For the reference population the dovitinib and sorafenib arms were combined (N=148). This allowed the calculation of a percentile score for each patient. Patients were grouped according to RECIST v1.1 response (centrally assessed) and included in FIG. 1. As is seen in FIG. 1, the higher the percentile score of a patient, the higher is the predicted likelihood of response to the treatment.
example 3
in Patients with Gastrointestinal Stromal Tumor Refractory and / or Intolerant to Imatinib (ClinicalTrials.Gov Identifier: NCT01478373)
[0405]Gene expression data (Affymetrix) and clinical response data was obtained from 14 patients. The DRP was applied to calculate a score for each patient (mean of genes in Table 2 minus mean of genes in Table 3). The score for each patient was compared to a reference population with the same diagnosis. For the reference population public data sets were downloaded from the GEO database (GSE20708, GSE17743, GSE8167, N=83). This allowed the calculation of a percentile score for each patient. Patients were grouped according to RECIST v1.1 response (locally assessed, no central assessment) and included in FIG. 1. As is seen in FIG. 1, the higher the percentile score of a patient, the higher is the predicted likelihood of response to the treatment.
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com