Nitrilase from arabis alpina, its encoding gene, vector, recombinant bacterial strain and uses thereof
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0027]Preparation of Nitrilase Aa-Nit
[0028](1) Nitrilase Aa-Nit amino acid sequence and nucleic acid sequence. A nitrilase amino acid sequence (Genbank No. KFK44999.1) was obtained via screening nitrilase gene sequence from protein database PDB and NCBI. The nitrilase comes from Arabis alpina, a plant belong to genus Arabis, family Brassicaceae. Based on the amino acid sequence of nitrilase, optimized codons from E. coli preferred codons, and the characteristics of vector pET28b(+), restriction enzyme cutting sites Xho I and Xba I were selected. The nitrilase-coding nucleic acid (shown in SEQ ID No. 2) and the coded amino acid sequence (shown in SEQ ID No. 1) were synthesized.
[0029](2) Construction of recombinant strain. The nucleic acid segment was treated with restriction endonucleases Xho I and Xba I and recovered. The recovered gene and commercial vector pET28b(+) (pre-treated with restriction endonucleases Xho I and Xba I) were treated with T4 DNA ligase for 16 hr at 16° C. to ...
example 2
[0031]Catalysis with Nitrilase Aa-Nit-Containing Resting Cells
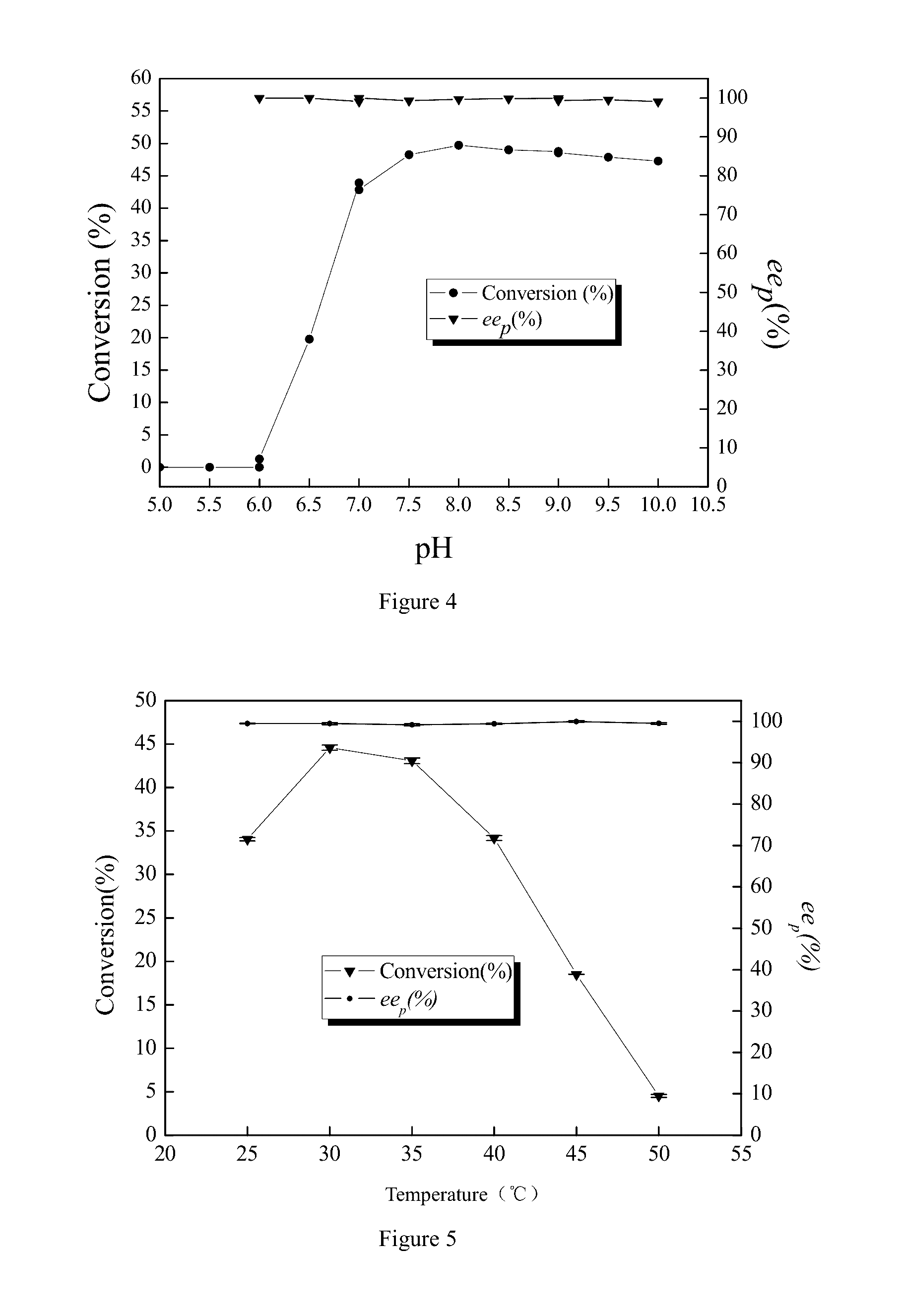
[0032]The optimal pH, temperature, pH stability and substrate tolerance were investigated.
[0033]Reaction mixture (10 mL) was composed of buffer solution (10 mL, buffer), racemic IBSN (substrate), and wet resting cells (catalyst). The substrate's concentration was 0.4 mol / L. The catalyst quantity was 20 g of wet resting cells / L. The resting cells contained 70-90% of water. The reaction was initiated in a water bath shaker at 150 rpm for 0.5 hr and terminated with 2M HCl. The conversion rate was obtained with gas chromatography to ascertain the catalytic activity of the resting cells under various conditions.
[0034](1) Determination of optimal pH. With the catalysis system defined above, the conversion rate of racemic IBSN was determined under various pH values (pH=5, 5.5, 6, 6.5, 7, 7.5, 8, 8.5, 9, 9.5, and 10) at 30° C. with substrate concentration at 0.4 mol / L and wet resting cells at 20 g / L. The buffers for various pH we...
example 3
[0038]Application of Nitrilase Aa-Nit-Containing Resting Cells
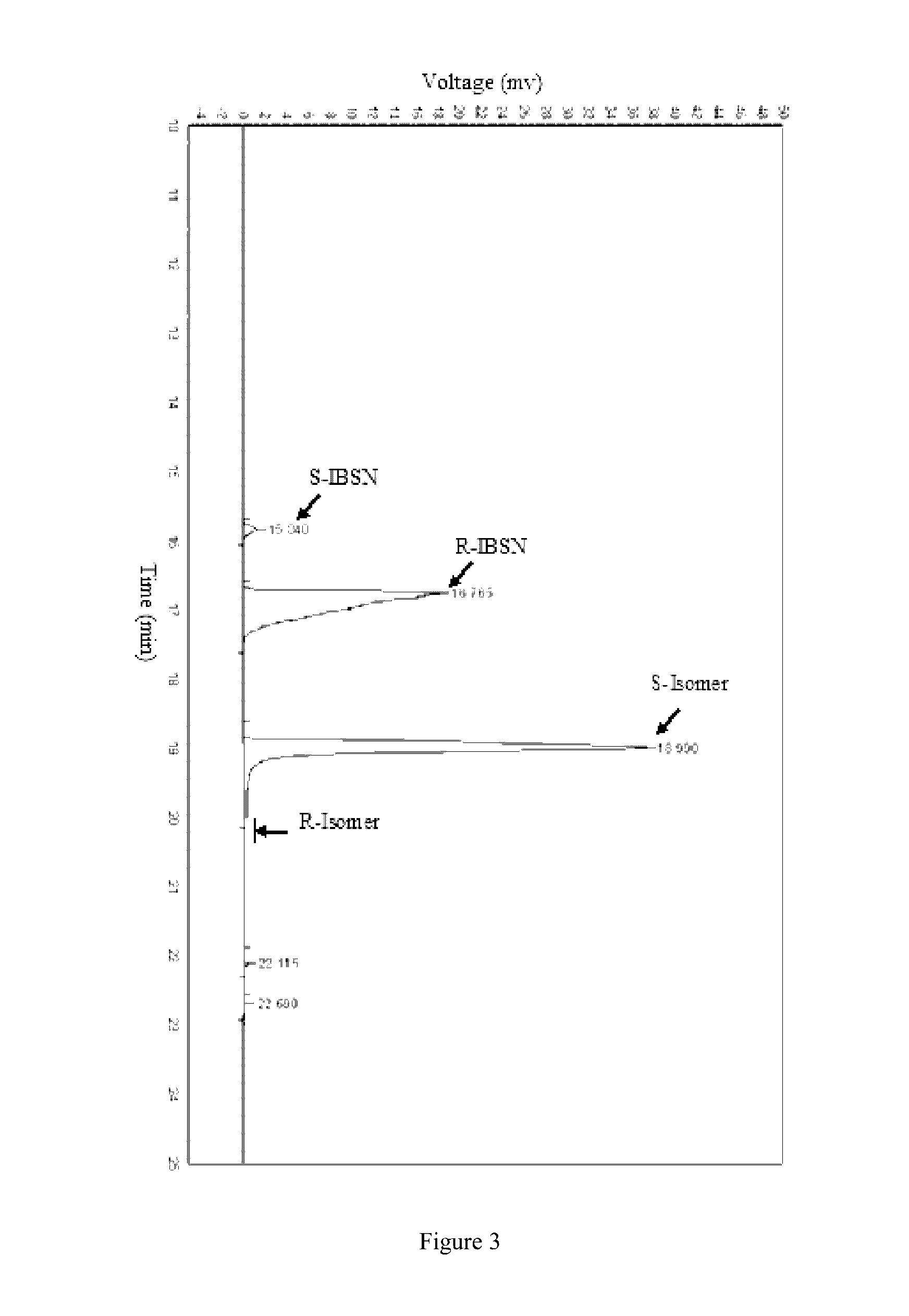
[0039]Kinetic resolution of racemic IBSN. The reaction is shown in FIG. 1. To a Tris-HCl buffer (10 mL, 100 mM, pH 8.0) was added racemic IBSN to reach 1.2 M, and 0.5 g of wet resting cells prepared in Example 1. The mixture was shaken at 30° C. for 15 hr in a water bath shaker. A 500 μL sample was taken every 3 hr and the reaction was quenched with 200 μL of 2 M HCl. The mixture was extracted with 800 μL of ethyl acetate, shaken, and centrifuged (12000× g, 2 min). The supernatant was dried with anhydrous sodium sulfate and analyzed with gas chromatography.
[0040](1) Determination of conversion rate and ee value with chiral gas chromatography. The amount of substrate and product present in the extract was determined with chiral gas chromatography (GC-14 C, Shimadzu, Japan). The capillary tube was BGB-174 (BGB Analytik, Switzerland). Gas chromatography conditions are below:
Sample amount1 μLInlet and detector temperature220°...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com