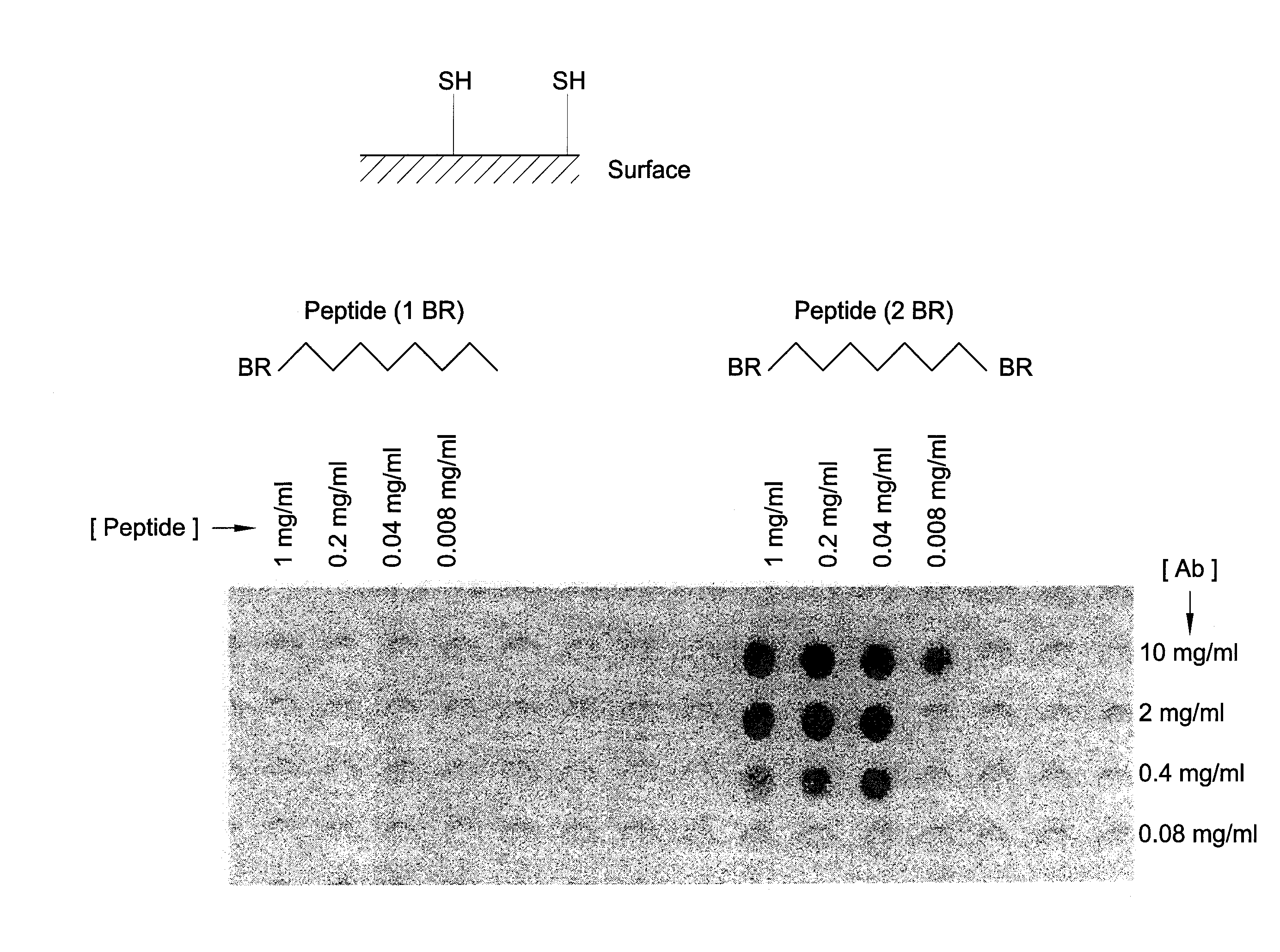
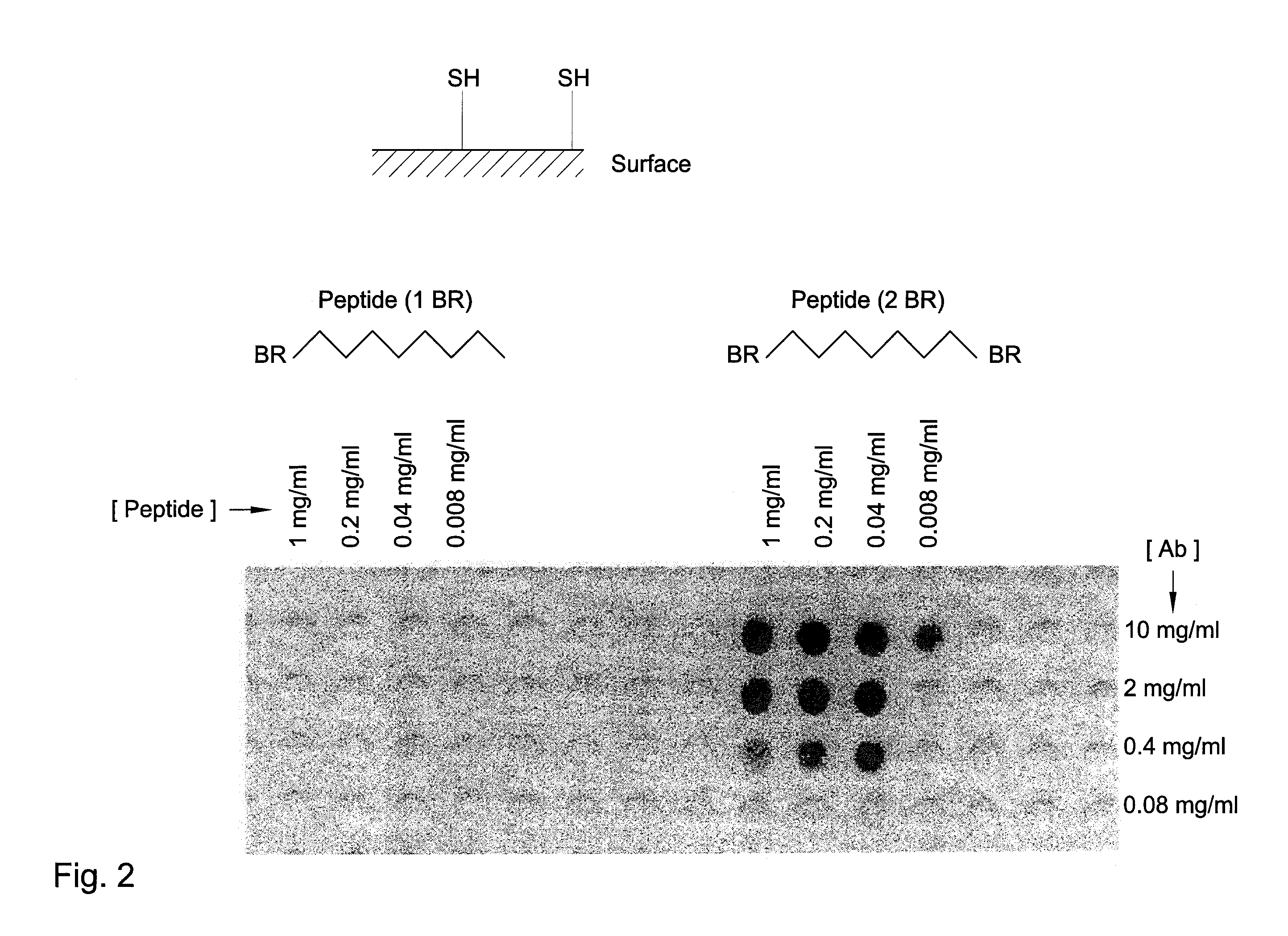
Identification of protein binding sites
a protein and site technology, applied in the field of molecular recognition or detection, can solve the problem of not paying attention to synthesizing specific segments in close proximity, and achieve the effect of quick and straightforward identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0082]A polypropylene or polyethylene support, or of other suitable material, was grafted with, for instance, polyacrylic acid. As an example: a polypropylene support in a 6% acrylic acid solution in water containing CuSO4 was irradiated using gamma radiation at a dose of 12 kGy. The grafted solid support containing carboxylic acid groups was functionalized with amino groups via coupling of t-butyloxycarbonyl-hexamethylenediamine (Boc-HMDA) using dicyclohexylcarbodiimide (DCC) with N-hydroxybenzotriazole (HOBt) and subsequent cleavage of the Boc groups using trifluoroacetic acid. Subsequently, the surface is functionalized with (when preferred, a mixture of differently protected) Cys amino acids using standard Fmoc chemistry. Examples of differently protected Cys groups are Cys (Trt) and Cys (mmt). After removal of the FMOC, the amino group is acetylated. Side chain deprotection can be done as described. Standard Fmoc peptide synthesis chemistry was used to link peptides (segments) ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| distances | aaaaa | aaaaa |
| distances | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com