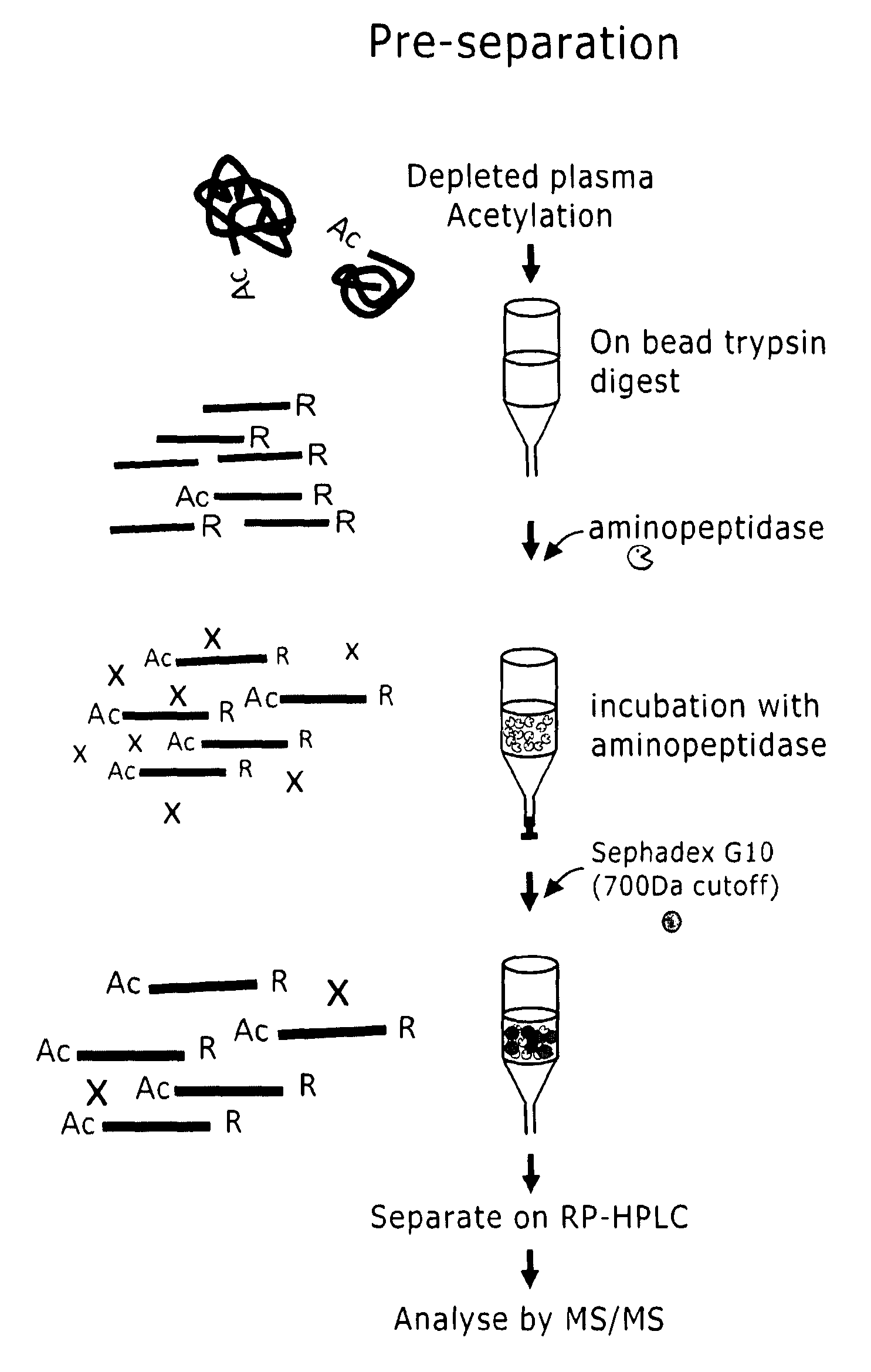
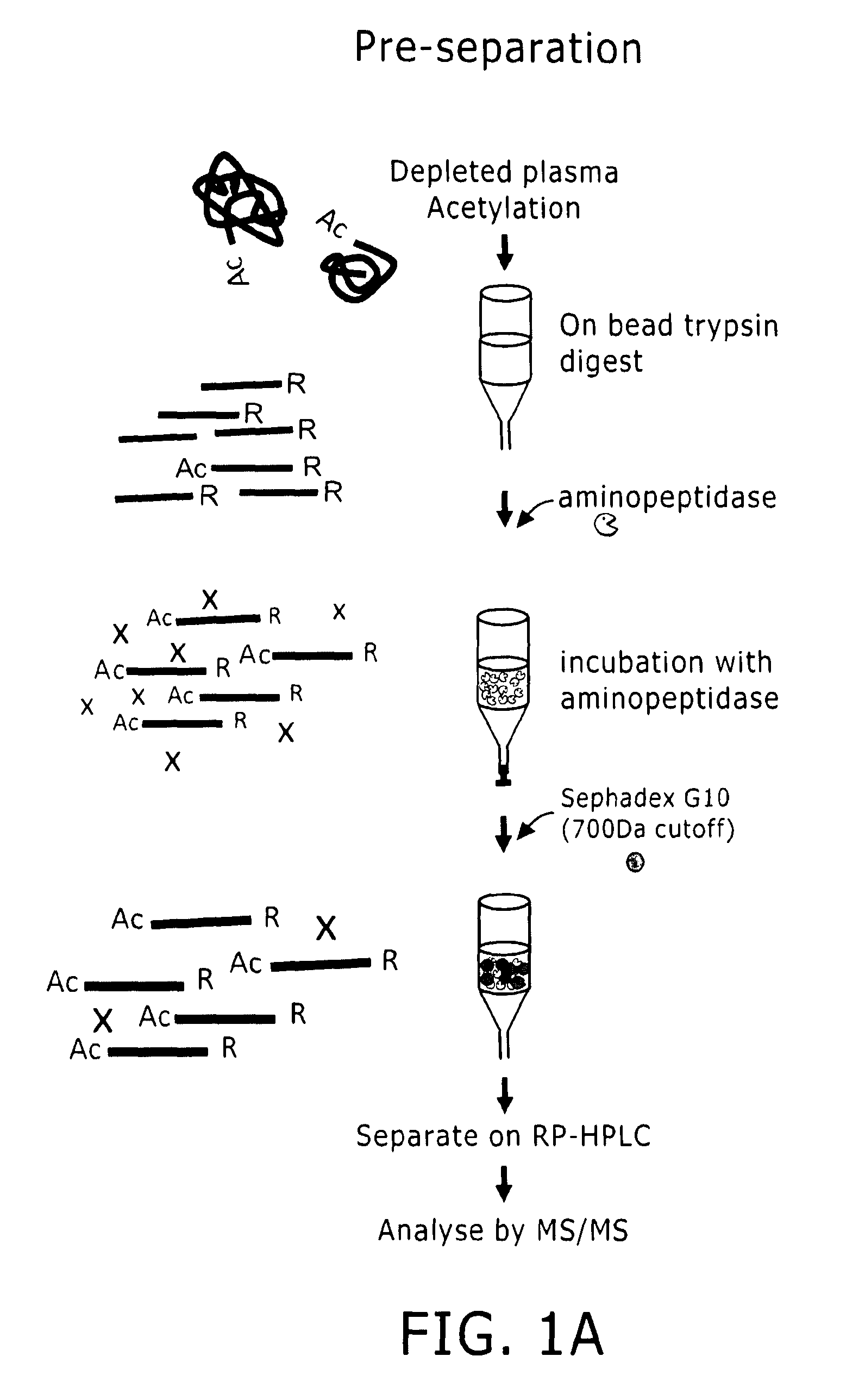
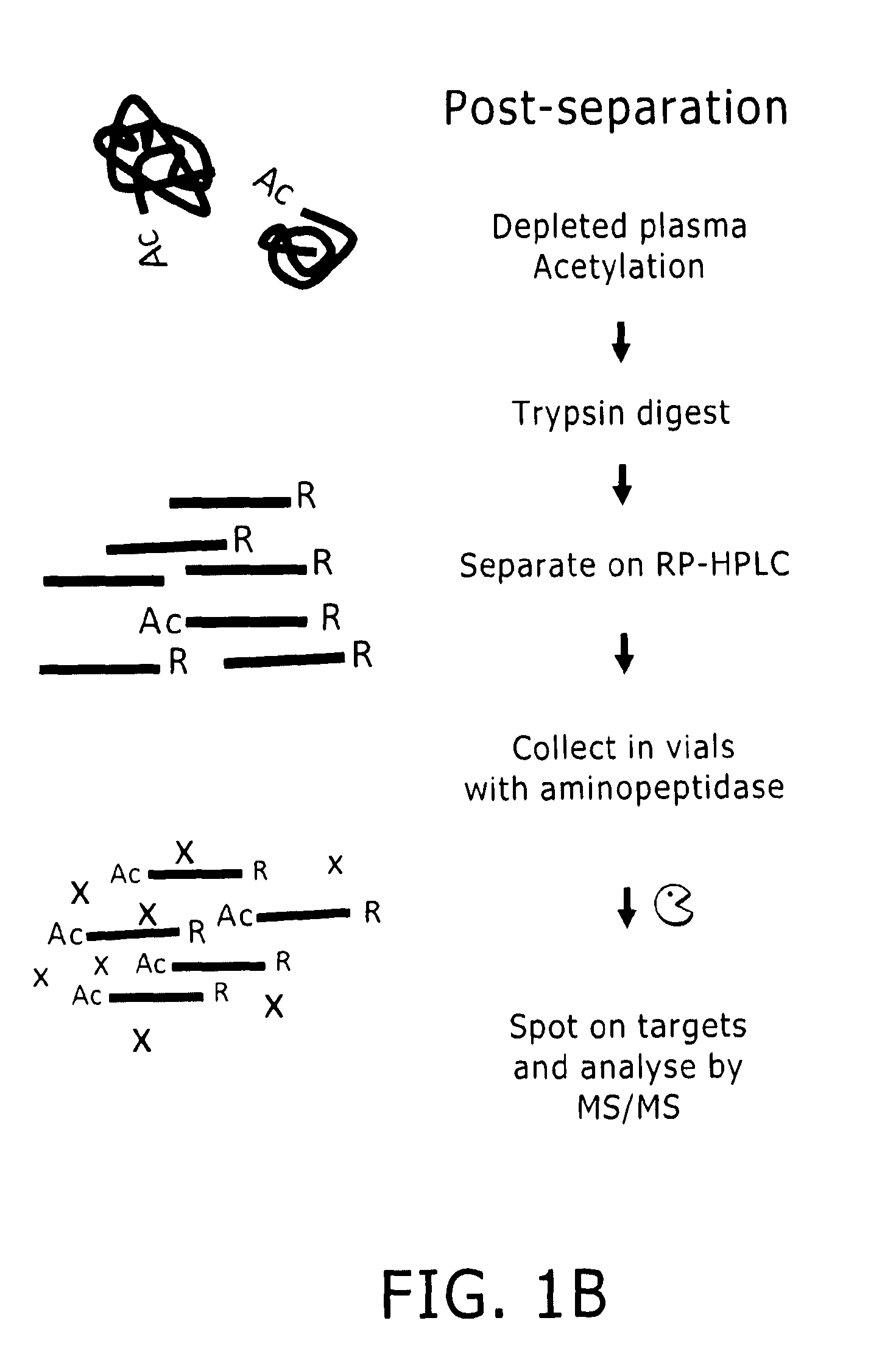
Preparation of samples for proteome analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Acetylation Protects Peptides from Degradation by Aminopeptidase
[0135]In the first experiment we investigated the effect of using acetylation as N-terminal blocking on progressive hydrolysis of peptides by bacterial aminopeptidase. As substrate for aminopeptidase activity we used the unprotected peptides from the Pepmix4 calibration mix (PepMix4, LaserBioLabs #C104). One tube was dissolved in 100 μl water resulting in peptide concentrations ranging form 8-50 pmoles / μl. As protected peptides we used 2 acetylated peptides designated P1380 and P1384 (for sequences see table 1). Both were diluted to 40 pmoles / μl. The sample reaction mixture that was used contained 1 μl of PepMix4 and 1 μl of each P1380 and P1384, all added to 20 mM TrisHCl buffer pH8, to a final volume of 20 μl. One sample was incubated overnight at 37° C. with 1 unit of Aeromonas proteolytica aminopeptidase (Sigma-aldrich, A8200) while another sample was left untreated. The reaction was stopped with 10% Trifluoroacetic...
example 2
ICPL Leads to N-terminal Protection from Aminopeptidase Activity
[0136]We investigated whether other blocking groups could improve the protection from aminopeptidase activity. In this experiment we also studied the use of Aminopeptidase M purified form pig kidney microsomes.
[0137]To prevent contamination issues with the commercial aminopeptidase M preparation (Sigma Aldrich, #L5006), the enzyme batch was dialyzed against 60 mM Sodiumphosphate at pH7. The ICPL reagent (Serva ICPL™-kit, #39230) was employed as a blocking agent to improve the protection from aminopeptidase activity. The rationale behind this is the greater sterical hindrance for enzyme activity if a bigger group is introduced on the N-terminus. ICPL labeling of 4 peptides designated L2 to L5 (Table 1), was performed according to the manufacturer's instructions. A mixture was prepared with 200 pmoles of the Pepmix4 peptides, 50 pmoles of each L2 to L5 and 0.05U of the dialyzed aminopeptidase M. An overnight incubation at...
example 3
The Modified N-terminal Peptides of Proteins are more Resistant to Aminopeptidase Activity when Compared to their Internal Peptides
[0138]This experiment was performed to show that the N-terminal peptides can be enriched by their altered susceptibility to aminopeptidase activity.
[0139]For this analysis, we prepared a mixture of 7 proteins (α-1-antitrypsin, hemoglobin, transferrin, albumin, β-lactoglobulin, α-1-acid glycoprotein and catalase; all from Sigma-aldrich) in PBS containing 4M Guanidine HCl. A total of 0.7 mg protein (0.1 mg / protein) was used for sample preparation. The sample was first reduced by treatment for 10′ at 30° C. with 6 μl 0.1M Tris(2-carboxyethyl)-phosphine Hydrochloride (TCEP.HCl, Pierce, #20490) after adjusting to pH 7.0. After this reduction, the free sulfhydryl groups on cysteines are alkylated by adding 6 μl 0.2M iodoacetamide (Fluka, #57670) for 60′ at 30° C. The sample was subsequently brought on a NAP™5 gelfiltration column (GE healthcare, #17-0853-02) t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Mass | aaaaa | aaaaa |
| Mass | aaaaa | aaaaa |
| Mass | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com