In Vivo Site-Specific Incorporation of N-Acetyl-Galactosamine Amino Acids in Eubacteria
a technology of nacetylgalactosamine and amino acids, applied in the field of protein biochemistry, can solve the problems of difficult analysis of glycan structure and study of glycosylation effects on protein structure and function, and the method can become problemati
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
System for Incorporating a Glycosylated Amino Acid into Proteins
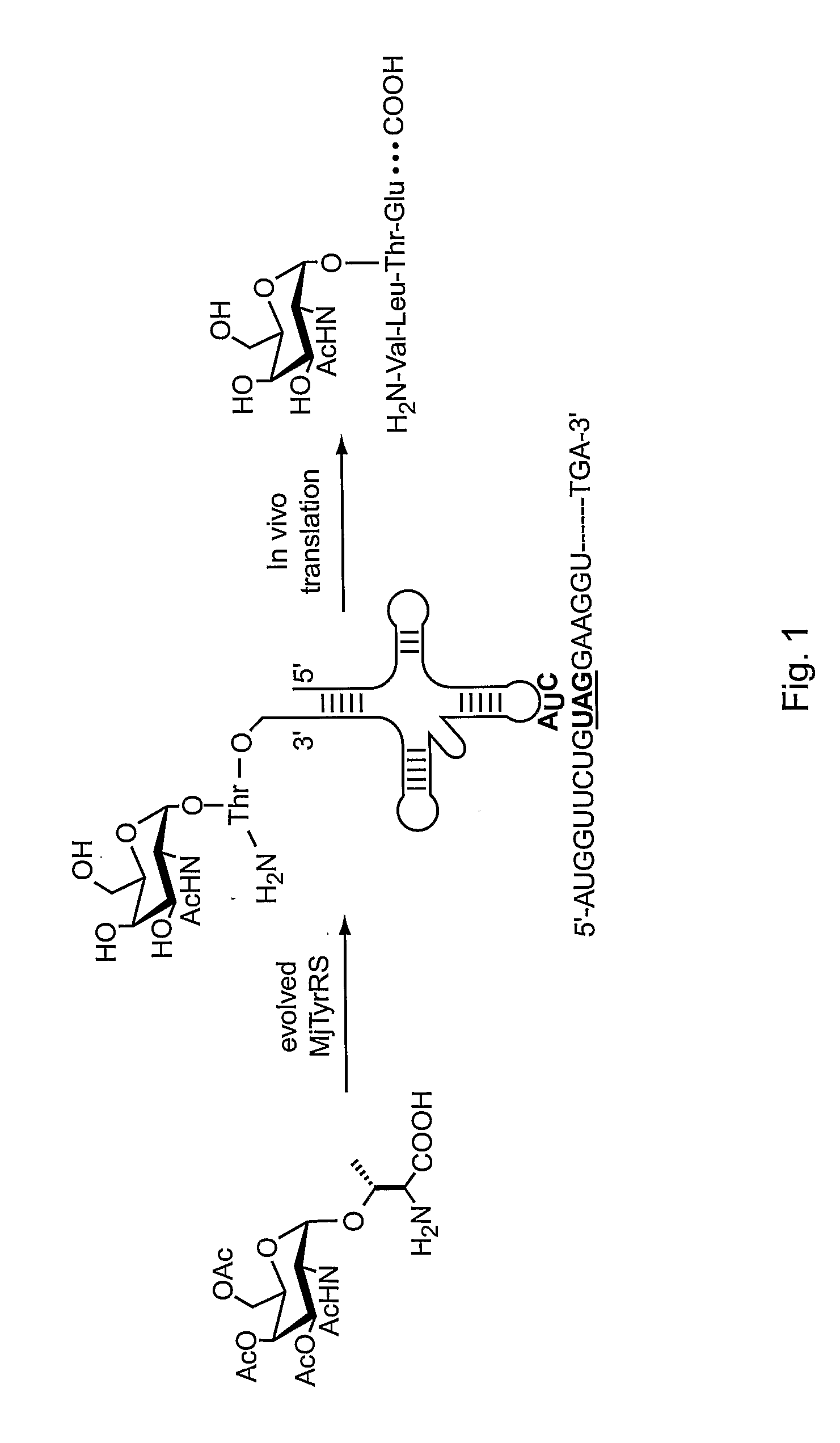
[0294] In eukaryotes, mucins comprise the most prevalent type of O-glycans, constituting a polydisperse group of glycoproteins and proteoglycans involved in inflammation and cellular recognition (Hanisch (2001) Biol. Chem. 382:143-149). The N-acetylgalacosamine (GalNAc) saccharide α-linked to the hydroxyl group of threonine or serine represents the core unit (the “Tn antigen”) in mucin-type glycoproteins. A wide variety of mucin-type core structures are generated by subsequent glycosylation of the GalNAc residue at the C-3 and / or C-6 hydroxyl groups.
[0295] This example describes methods and compositions for preparing N-acetylgalactosamine-α-threonine (GalNAc-Thr) and co-translationally incorporating this unnatural amino acid into a protein in vivo. In order to genetically encode this functional group in E. coli in the form of GalNAc-threonine, a number of tRNA-synthetase pairs were evolved that are capable of insertin...
example 2
Strategy for the Synthesis of Glycoproteins
[0314] In another embodiment of the invention, homogeneous glycoproteins are synthesized in an organism, e.g., E. coli, by the cotranslational incorporation of the glycosylated amino acid N-acetylglucosamine-β-serine (GlcNAc-Ser). For example, myoglobin containing β-GlcNAc-serine at a defined position can be expressed in E. coli in good yield and with high fidelity. The β-GlcNAc moiety can be recognized by a carbohydrate binding protein or subsequently modified with a galactosyltransferase. This approach can also be applicable to other posttranslational modifications, e.g., protein phosphorylation, acetylation, methylation and the like.
[0315] Methods were previously developed which for the first time allowed the systematic addition of amino acids with novel chemical and physical properties to the genetic code of E. coli (see, e.g., L. Wang, et al., (2001) Science 292:498; L. Wang, et al., (2002) J. Am. Chem. Soc. 124:1836; Z. Zhang, et al...
example 3
Sequences of Exemplary O-RSs
[0328] Exemplary O-RSs that can be used in the invention include SEQ ID NOS.: 1-4 and 11-13 (See Table 3), and exemplary O-tRNA that can be used in the invention includes SEQ ID NO: 17. Exemplary polynucleotides that encode O-RSs include SEQ ID NOS.: 6-9 and 14-16.
[0329] It is understood that the examples and embodiments described herein are for illustrative purposes only and that various modifications or changes in light thereof will be suggested to persons skilled in the art and are to be included within the spirit and purview of this application and scope of the appended claims.
[0330] While the foregoing invention has been described in some detail for purposes of clarity and understanding, it will be clear to one skilled in the art from a reading of this disclosure that various changes in form and detail can be made without departing from the true scope of the invention. For example, all the techniques and apparatus described above can be used in va...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Tm | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com