L-Amino Acid-Producing Bacterium and Method for Producing L-Amino Acid
a technology of l-amino acid and bacterium, which is applied in the field of l-amino acid-producing bacterium and method for producing lamino acid, can solve the problems that the use of methylophilus /i>bacteria in the production of l-amino acids has not been reported, and achieves enhanced l-amino acid biosynthetic enzyme activity, enhanced dihydrodipicolinate synthase activity and aspar
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Creation of L-Lysine-Producing Bacterium: (I)
[0165] (1) Introduction of Mutant lysC and Mutant dapA into Methylophilus Bacterium
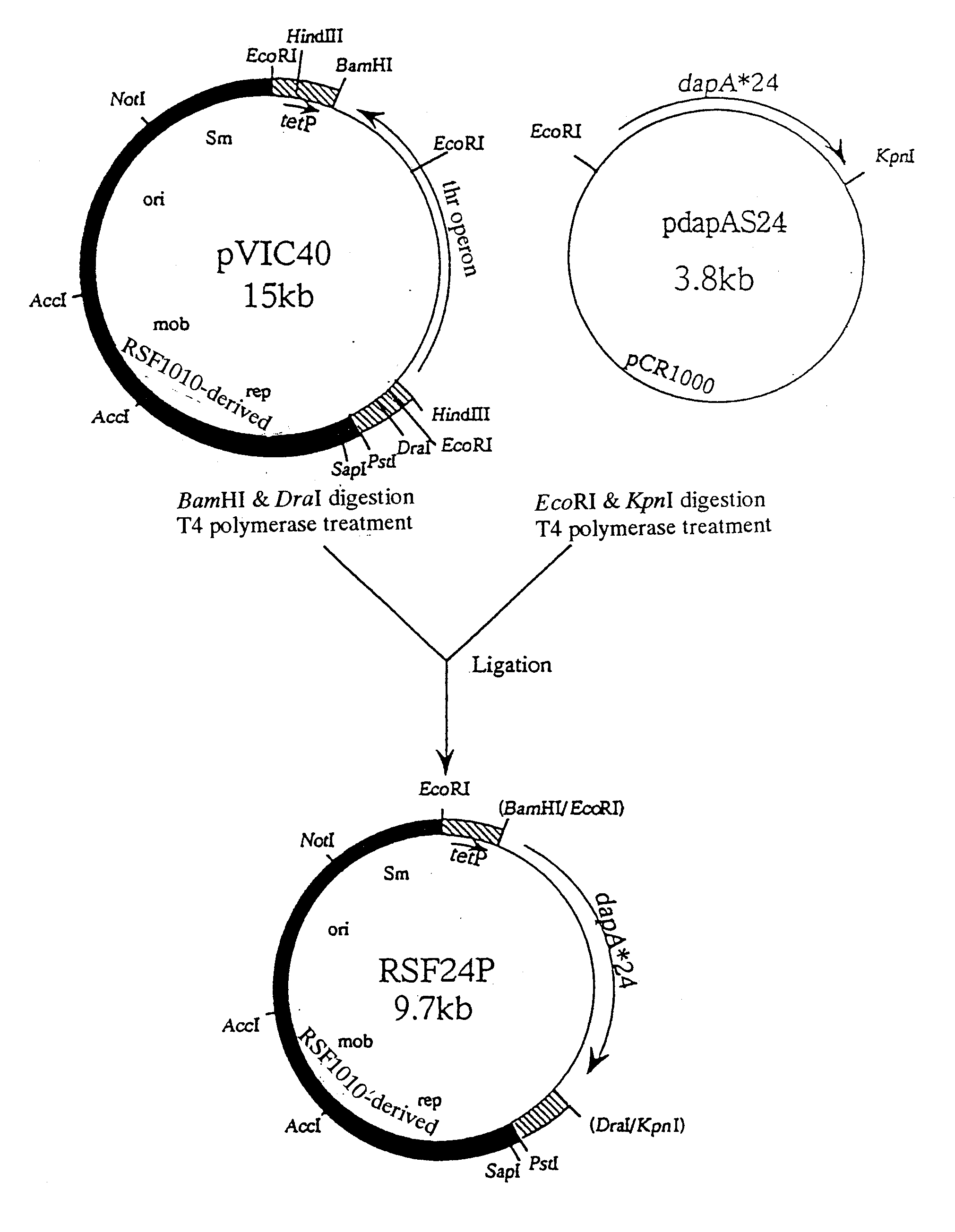
[0166] A Methylophilus bacterium was transformed with plasmid RSFD80 (see WO95 / 16042) which contained a mutant lysC and a mutant dapA. RSFD80 is plasmid pVIC40 (International Publication WO90 / 04636, Japanese Patent Application Laid-open (Kohyo) No. 3-501682 / 1991) derived from the broad host spectrum vector plasmid pAYC32 (Chistorerdov, A. Y., Tsygankov, Y. D., Plasmid, 16, 161-167, (1986)), which is a derivative of RSF1010. RSF1010 contains a mutant dapA and a mutant lysC derived from E. coli located downstream of the promoter (tetP) of the tetracycline resistance gene of pVIC40 in this order, so that the transcription directions of the genes are ordinary with respect to tetP. The mutant dapA codes for a mutant DDPS with a tyrosine in place of the histidine at position 118. The mutant lysC codes for a mutant AKIII with an isoleucine in place of the threon...
example 2
Creation of L-Lysine-Producing Bacterium (II)
[0180] (1) Introduction of the tac Promoter Region into a Broad Host Spectrum Vector
[0181] In order to produce a large amount of a L-lysine biosynthetic enzyme in Methylophilus methylotrophus, the tac promoter was used for gene expression of this target enzyme. This promoter is frequently used in E. coli.
[0182] The tac promoter region was obtained by amplification through PCR using pKK233-3 (Pharmacia) as the template, DNA fragments having the nucleotide sequences of SEQ ID NOS: 15 and 16 as primers, and a heat-resistant DNA polymerase. The PCR was performed with cycles of 94° C. for 20 seconds, 60° C. for 30 seconds, and 72° C. for 60 seconds, repeated 30 times. Then, the amplified DNA fragment was collected and treated with restriction enzymes EcoRI and PstI. The broad host spectrum vector pRS (see Japanese Patent Application Laid-open (Kohyo) No. 3-501682 / 1991) was also digested with the same restriction enzymes, and the aforementio...
example 3
Creation of L-Lysine-Producing Bacterium (III)
[0189] The Methylophilus methylotrophus AS1 strain (NCIMB10515) was inoculated into 121M1 medium and cultured at 37° C. for 15 hours. The obtained bacterial cells were treated with NTG in a conventional manner (NTG concentration: 100 mg / L, 37° C., 5 minutes), and spread onto 121M1 agar medium containing 7 g / L of S-(2-aminoethyl)-cysteine (AEC) and 3 g / L of L-threonine. The cells were cultured at 37° C. for 2 to 8 days, and the colonies which formed were picked up to obtain AEC-resistant strains.
[0190] The aforementioned AEC-resistant strains were inoculated into 121 production medium, and cultured at 37° C. for 38 hours under aerobic conditions. After the culture was completed, the cells and calcium carbonate were removed from the medium by centrifugation, and the L-lysine concentration in the culture supernatant was measured by an amino acid analyzer (JASCO Corporation [Nihon Bunko], high performance liquid chromatography). The strain...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com