Methods of detecting nucleic acids in individual cells and of identifying rare cells from large heterogeneous cell populations
a nucleic acid and individual cell technology, applied in the field of nucleic acid chemistry and biochemical assays, can solve the problems of insufficient progress in the validation of the clinical utility of ctc detection as a prognostic indicator, low concentration of ctc in peripheral blood or other body fluids, etc., and achieve the effect of improving the specificity of the assay
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
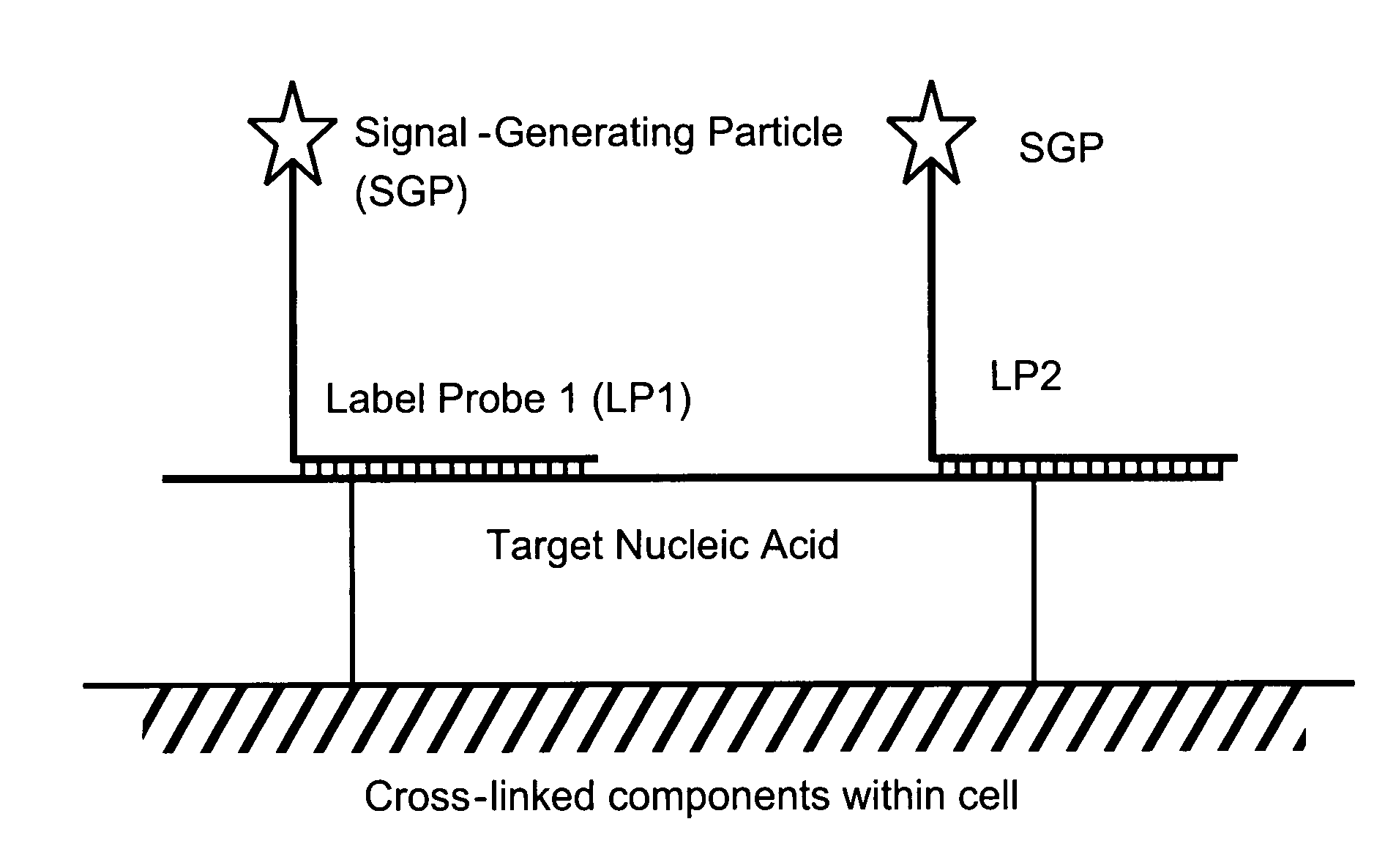
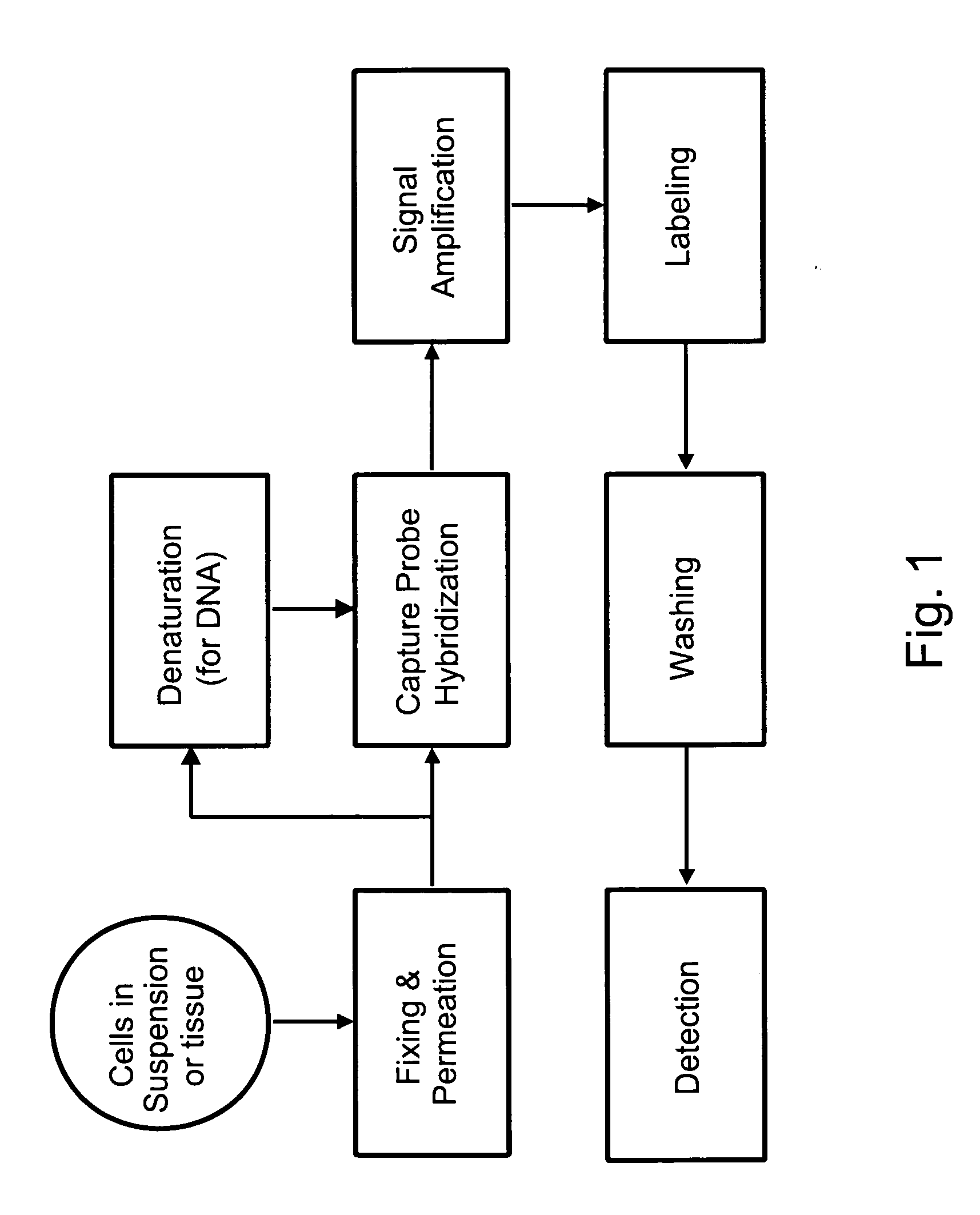
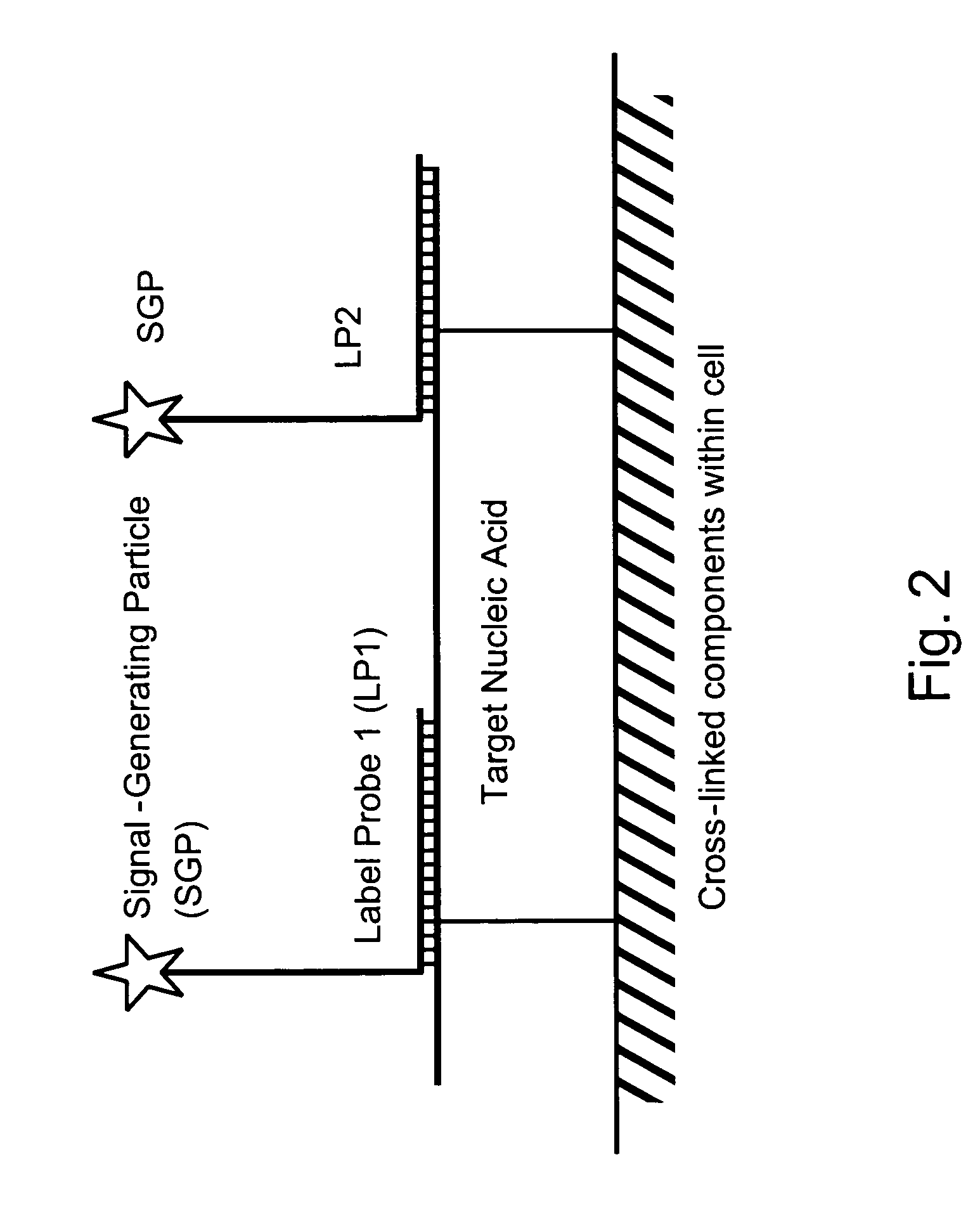
[0087]Among other aspects, the present invention provides multiplex assays that can be used for simultaneous detection, and optionally quantitation, of two or more nucleic acid targets in a single cell. A related aspect of the invention provides methods for detecting the level of one or more target nucleic acids relative to that of a reference nucleic acid in an individual cell.
[0088]In general, in the assays of the invention, a label probe is captured to each target nucleic acid. The label probe can be captured to the target through direct binding of the label probe to the target. Preferably, however, the label probe is captured indirectly through binding to capture probes, amplifiers, and / or preamplifiers that bind to the target. Use of the optional amplifiers and preamplifiers facilitates capture of multiple copies of the label probe to the target, thus amplifying signal from the target without requiring enzymatic amplification of the target itself. Binding of the capture probes ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| nucleic acid | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| composition | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com