Scfvs in photosynthetic microbes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Materials and Methods
[0052] PCR was performed using AMPLITAQ® Gold DNA polymerase kit (APPLIED BIOSYSTEMS™, CA, USA) and a MASTERCYCLER® Gradient thermal cycler (EPPENDORF™, Hamburg, Germany). The PCR reaction was carried out in 100 μl of solution containing 10 μl Roche 10×PCR buffer II, 1 mM MgCl2, 100 μM dNTP, 0.25 μM each primer, 10 ng genomic DNA as template, and 5 U polymerase. The PCR amplification cycles consisted of denaturation at 94° C. for 45 sec (1 cycle), followed by 29 cycles of 94° C. 45 sec, 53° C. 45 sec and 72° C. 45 sec, the last extension was at 72° C. for 5 min. PCR products are resolved on 1.5% agarose gel stained with ethidium bromide and purified using CONCERT™ PCR purification kit (LIFETECHNOLOGIES™).
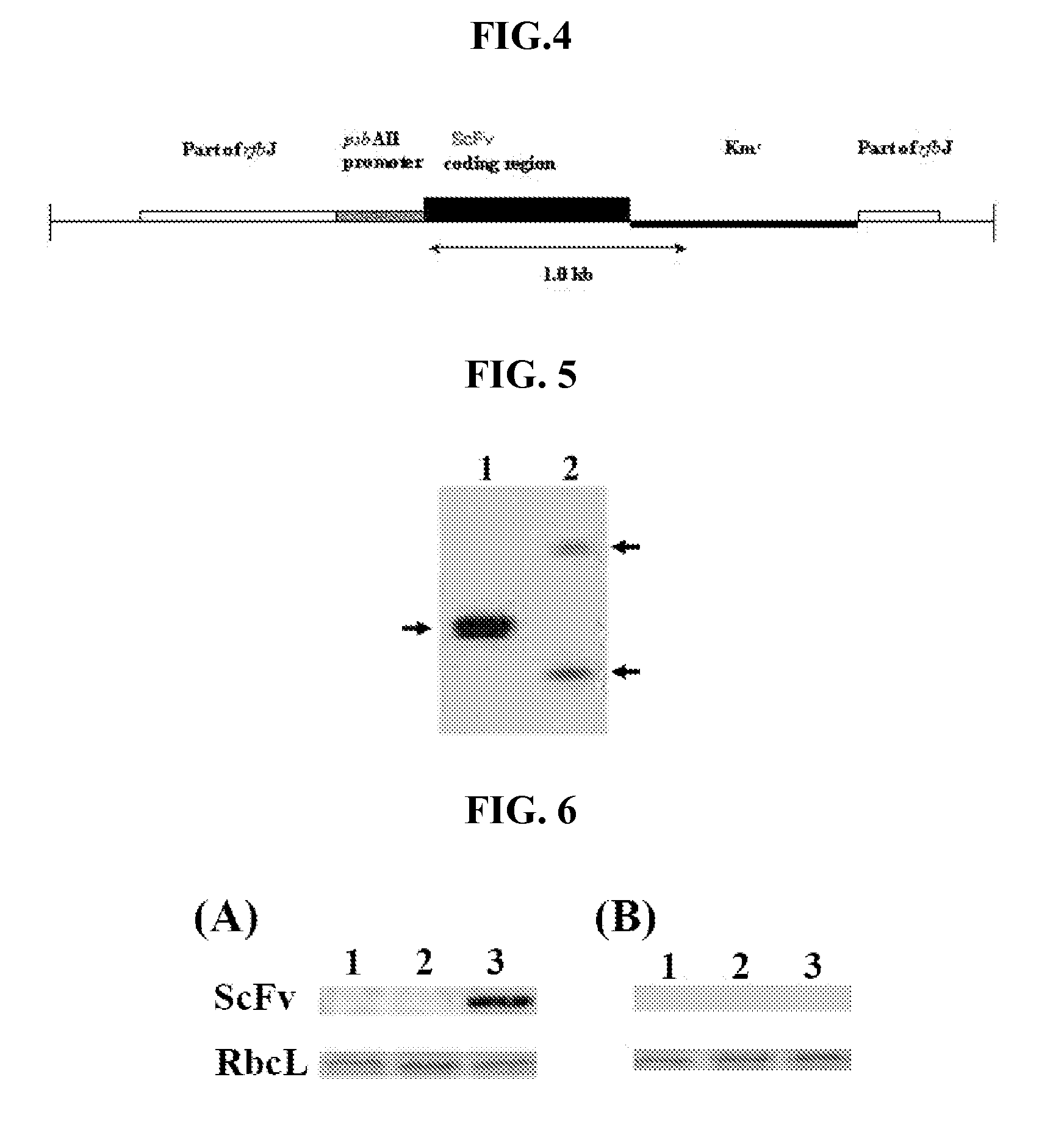
[0053] The recombinant DNA cassette illustrated in FIG. 4 was constructed by assembling within the coding region for rjbJ (SEQ ID NO: 7), the promoter region of Synechocystis PCC 6803 psbA II (SEQ ID NO: 10), a kanamycin resistance gene (KmR). The rjbJ coding ...
example 2
Expression of FcSv in Chlamydomonas
[0056]C. reinhardtii 950FcSv was generated by introducing a recombinant DNA cassette (FIG. 10) into the wild type C. reinhardtii. The box filled with crosshatch represents the coding sequence for ScFv with codons optimized for expression in C. reinhardtii, Ble is the bacterial bleomycin resistance gene serving as a dominant selectable marker (Stevens et al. 1996), Pro is the promoter region of RbcS2 in C. reinhardtii(Goldschmidt-Clermont and Rahire. 1986), Ω is the 5′untranslated leader (omega sequence) of tobacco mosaic virus (Schmitz et al. 1996), I is the first intron of RbcS2 (Lumbreras et al. 1998), C+K represent the ER retention sequence KDEL (SEQ ID NO: 14; Napier et al. 1992), T is the terminator of RbcS2 gene of C. reinhardtii. The coding sequence for ScFv was inserted using the NcoI site and NotI site.
[0057] In another embodiment, the present invention provides the details for the construction of a recombinant DNA molecule cassette for ...
example 3
Expression of ScFv-DB3 in Synechocystis
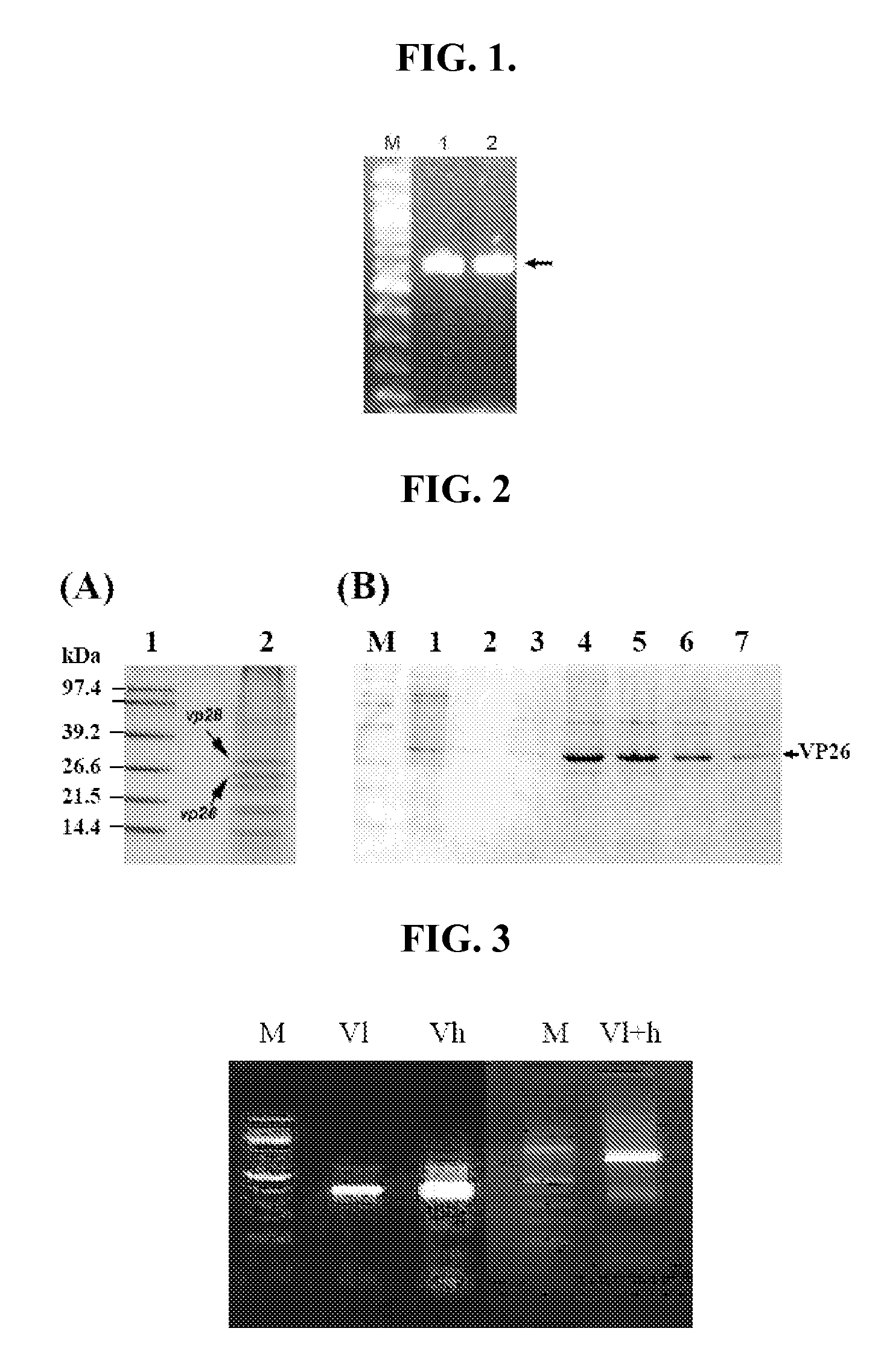
[0058] To investigate the production of ScFvs in cyanobacteria, the coding region for progesterone binding ScFvs DB3 was generated, incorporated into Synechocystis PCC 6803, and expressed in culture. Results indicate that functional DB3 was expressed in Synechocystis 6803 ScFvs DB3 and has a very high specificity for progesterone.
Synthesis of ScFvs DB3 Coding Sequence
[0059] An expression cassette for ScFvs DB3 in Synechocystis was designed which incorporated a 3′ flanking region of native genomic DNA, a promoter, an expression construct for the ScFvs DB3, a selection cassette, a termination sequence, and a 5′ flanking region of native genomic DNA. The ScFv-DB coding region (SEQ ID NO: 13) was synthesized based on the published sequence of DB3 (He et al. 1995) using the optimal codon bias for Synechocystis and incorporating an ATG start codon (instead of TGT). The coding region was inserted into the StuI restriction site of the recombinant D...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com