Methods for identifying small molecules that bind specific rna structural motifs
a technology of rna structural motifs and small molecules, applied in the field of screening and identifying test compounds, can solve problems such as difficult identification or design of synthetic agents, and achieve the effect of easy identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
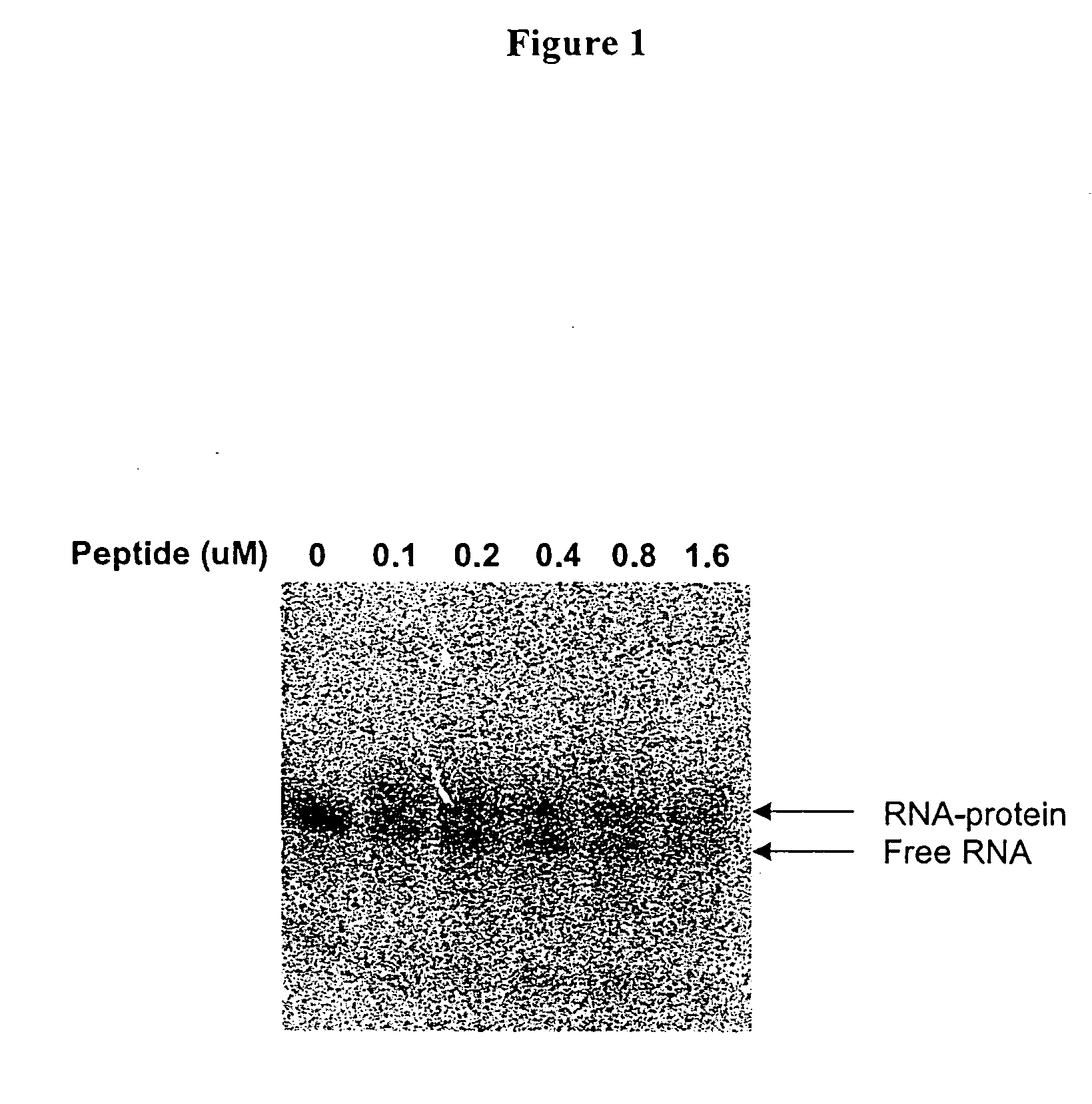
[0021] The present invention relates to methods for identifying compounds that bind to preselected target elements of nucleic acids, in particular, RNAs, including but not limited to preselected target RNA sequencing structural motifs, or structural elements. Methods are described in which a preselected target RNA having a detectable label is used to screen a library of test compounds. Any complexes formed between the target RNA and a member of the library are identified using physical methods that detect the altered physical property of the target RNA bound to a test compound. Changes in the physical property of the RNA-test compound complex relative to the target RNA or test compound can be measured by methods such as, but not limited to, methods that detect a change in mobility due to a change in mass, change in charge, or a change in thermostability. Such methods include, but are not limited to, electrophoresis, fluorescence spectroscopy, surface plasmon resonance, mass spectrom...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com