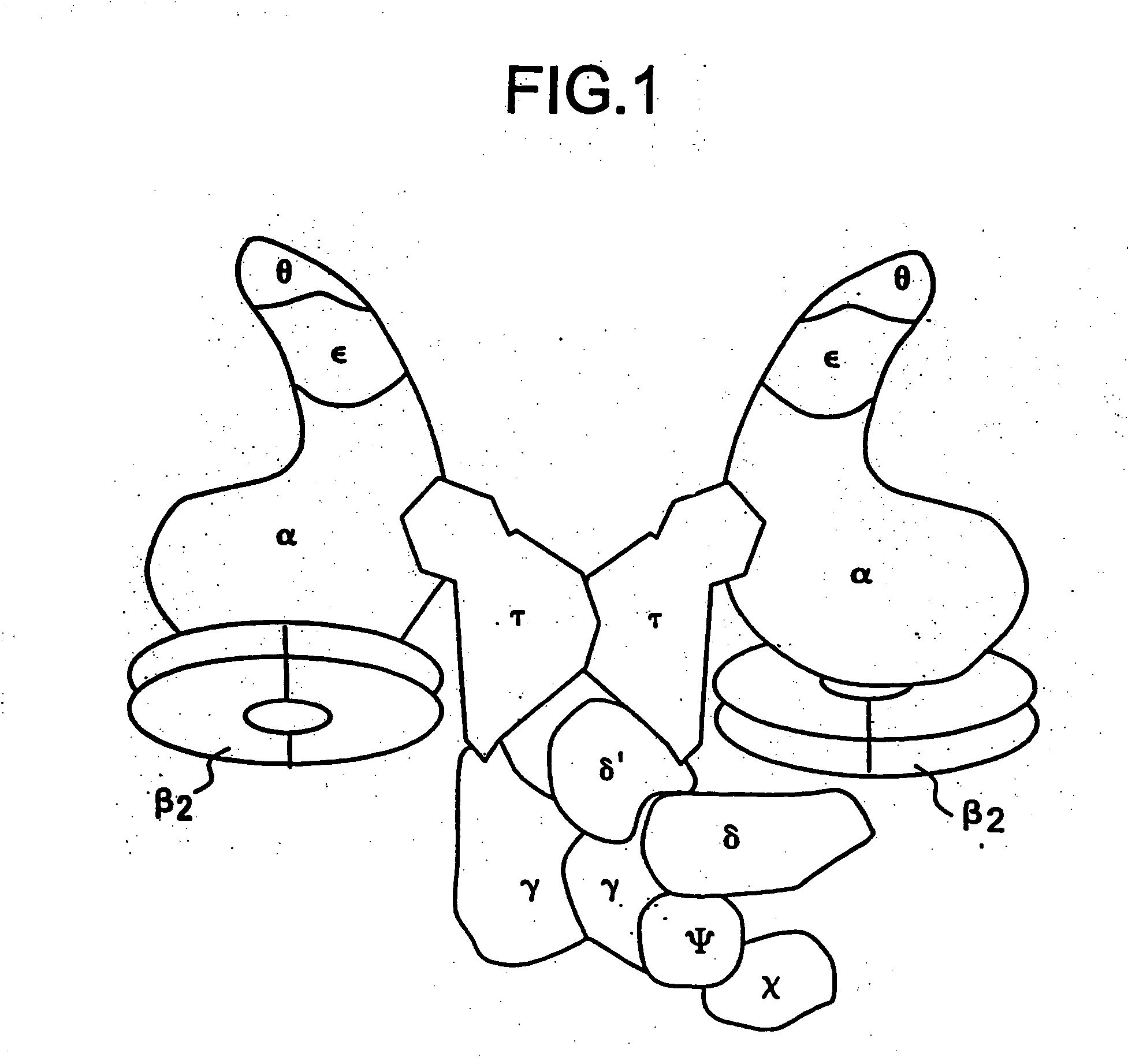
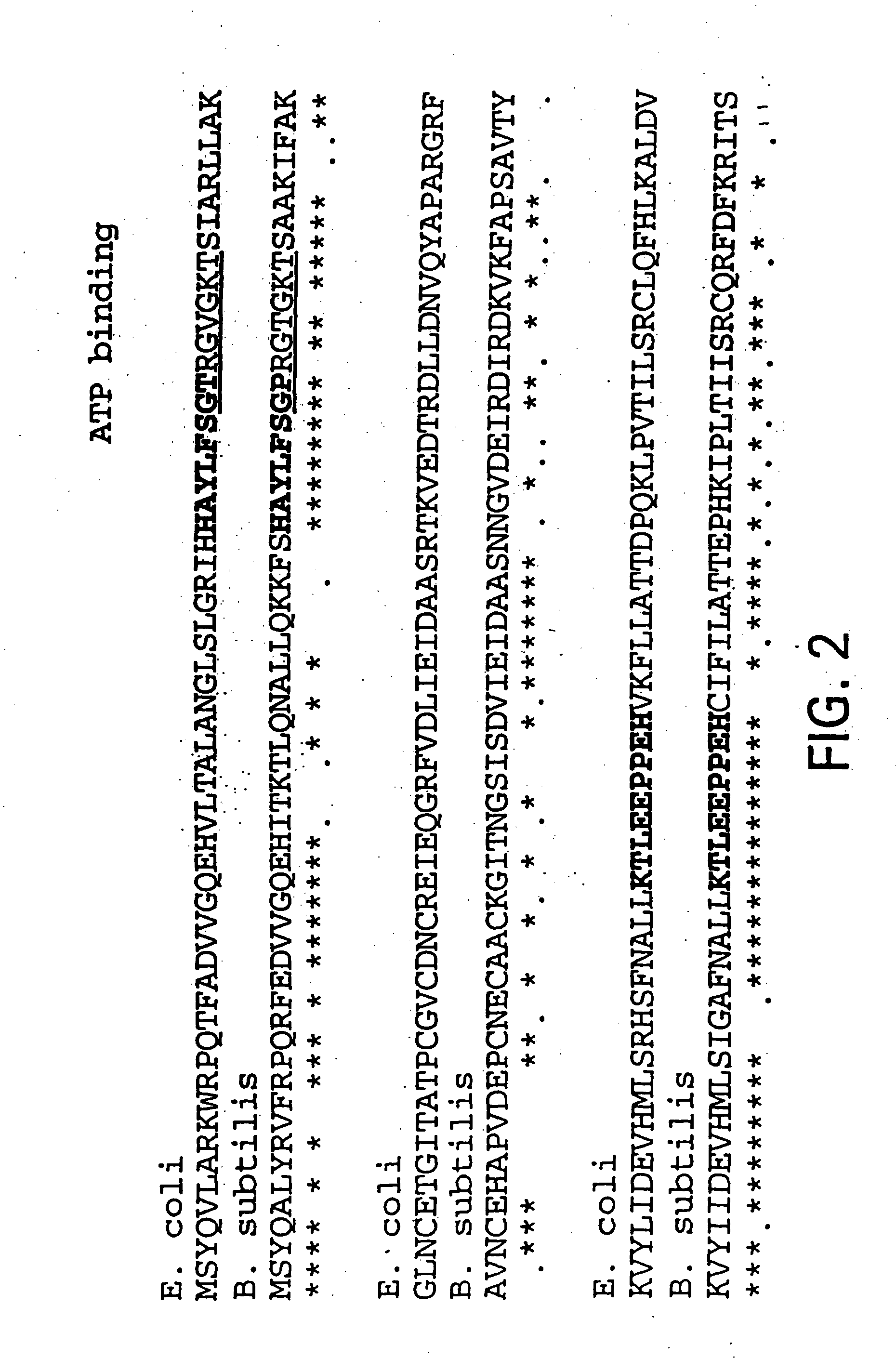
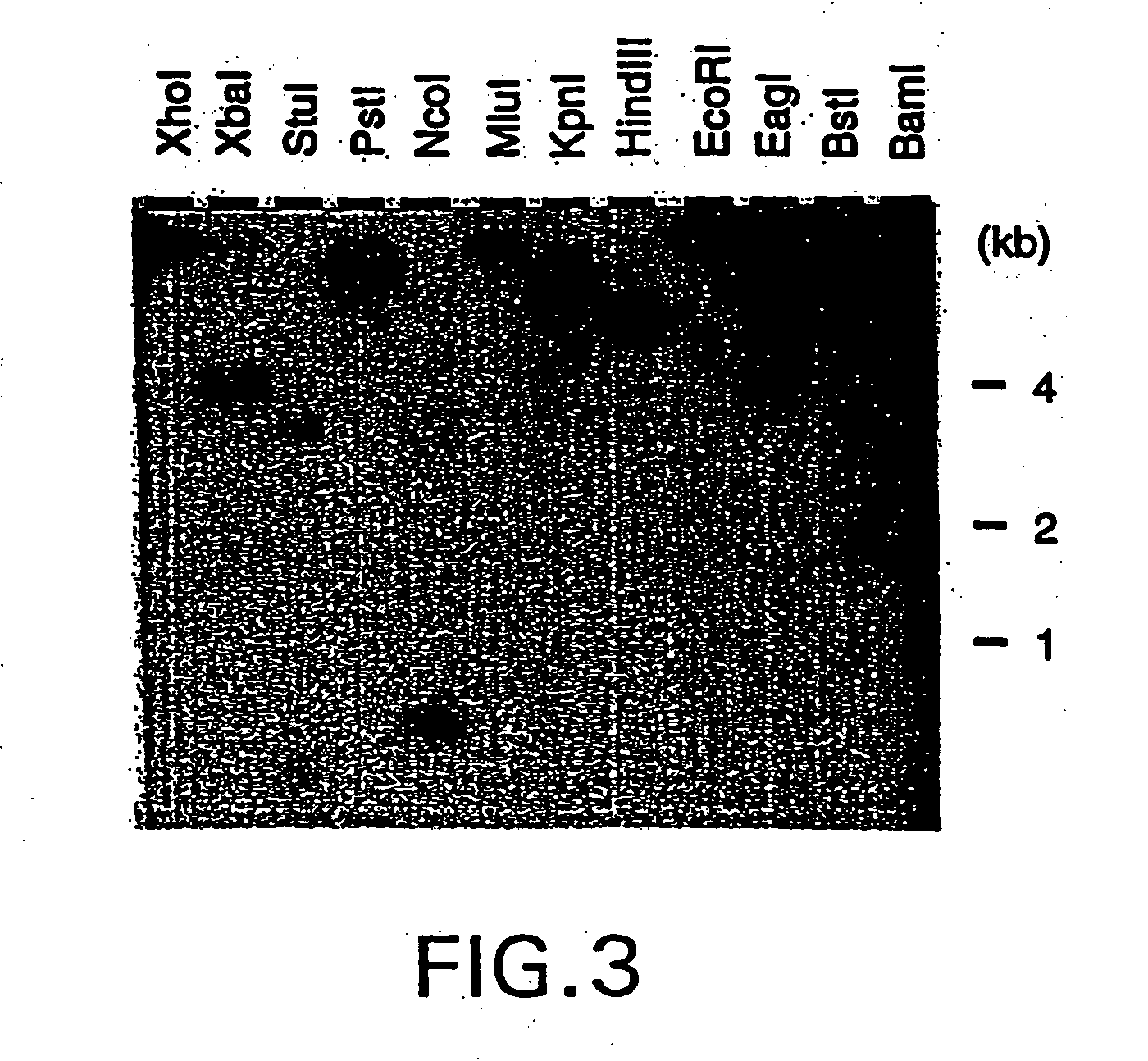
Nucleic acid encoding bacillus stearothermophilus tau polymerase subunit
a technology of bacillus stearothermophilus and polymerase, which is applied in the direction of enzymology, organic chemistry, transferases, etc., can solve the problems of lack of 3' to 5' exonuclease activity, limited thermostability of exant polymerases, and inability to obtain extended lengths of nucleotides
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0330] Experimental Procedures
[0331] Materials
[0332] DNA modification enzymes were from New England Biolabs. Labelled nucleotides were from Amersham, and unlabeled nucleotides were from New England Biolabs The Alter-1 vector was from Promega. pET plasmids and E. coli strains, BL21 (DE3) and BL21(DE3)pLysS were from Novagen. Oligonucleotides were from Operon. Buffer A is 20 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, 5 mMDTT, and 10% glycerol.
[0333] Genomic DNA
[0334] Thermus thermophilus (strain HB8) was obtained from the American Type Tissue Collection. Genomic DNA was prepared from cells grown in 0.1 l of Thermus medium N697 (ATCC: 4 g yeast extract, 8.0 g polypeptone (BBL; 11910), 2.0 g NaCl, 30.0 g agar, 1.0 L distilled water) at 75.degree. C. overnight. Cells were collected by centrifugation at 4.degree. C. and the cell pellet was resuspended in 25 ml of 100 mM Tris-HCl (pH 8.0), 0.05 M EDTA, 2 mg / ml lysozyme and incubated at room temperature for 10 min. Then 25 ml 0.10 M EDTA (pH 8.0), ...
example 2
[0364] Frameshifting Analysis of the T.th. dnaX Gene
[0365] Frameshifting was analyzed by inserting the frameshift site into lacZ in the three different reading frames, followed by plating on X-gal and scoring for blue or white colony formation (Weiss et al., 1987). The frameshifting region within T.th dnaX was subcloned into the EcoRI / BamHI sites of pUC19. These sites are within the polylinker inside of the .beta.-galactosidase gene. Three constructs were produced such that the insert was either in frame with the downstream coding sequence of .beta.-galactosidase, or were out of frame (either -1 or -2). An additional three constructs were designed by mutating the frameshift sequence and then placing this insert into the three reading frames of the .beta.-galactosidase gene. These six plasmids were constructed as described below.
[0366] The upstream primer for the shifty sequences was 5'-gcg cgg atc cgg agg gag aaa aaa aaa gcc tca gcc ca-3' (SEQ. ID. No. 10). The BamHI site for clonin...
example 3
[0369] Expression Vector for T.th. .gamma. and .tau.
[0370] The dnaX gene was cloned into the pET16 expression vector in the steps shown in FIG. 9. First, the bulk of the gene was cloned into pET16 by removing the PmlI / XbaI fragment from pAlterdnaX, and placing it into SmaI / XbaI digested Puc19 to yield Puc19dnaXCterm. The N-terminal sequence of the dnaX gene was then reconstructed to position an NdeI site at the N-terminus. This was performed by amplifying the 5' region encoding the N-terminal section of .gamma. / .tau. using an upstream primer containing an NdeI site that hybridizes to the dnaX gene at the initiating gtg codon (i.e. to encode Met where the Met is created by the PCR primer, and the Val is the initiating gtg start codon of dnaX). The primer sequence for this 5' end was: 5'-gtggtgcatatg gtg agc gcc ctc tac cgc c-3' (SEQ. ID. No. 15) (where the NdeI site is underlined, and the coding sequence of dnaX follows). The downstream primer hybridizes past the PmlI site at nucleot...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com