Rodent expression systems utilising polyoma virus and epstein barr virus sequences
A technique for rodents, expression systems, applied in the field of protein expression systems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Materials and methods:
[0088] cells and media
[0089] CHO-K1 (ATCCCCL 61) was grown in DMEM:COONS F12 containing 10% FBS. CHO-T cells were adapted for suspension growth in serum-free medium (Excell302; JRH Biosciences, Kansas) and protein-free medium (Excell325; JRH Biosciences, Kansas). XL99 cells (a suspension CHO-K1 cell line suitable for suspension growth in serum free medium (Neil Kitchen 1999)) were grown in Excell302.
[0090] NS0-T
[0091] Plasmids encoding the polyoma virus large T antigen were transfected with NSO cells (non-secreting mouse myeloma cells) (ECACC 85110503).
[0092] Electroporation of NS0
[0093] Electroporation of NSO cells was performed using the appropriate protocol described by Chu G et al. 1987. In short, put 1-1.5 x 10 7 NS0 cells were grown to mid-log phase and electroporated with 10 μg pBK-CMV-large T antigen plasmid at 250V with 15 ms square wave pulses. Cells were selected in G418 (400 μg / ml) 48 hours after transfection. ...
Embodiment 2
[0124] Episomal replication in CHO cells
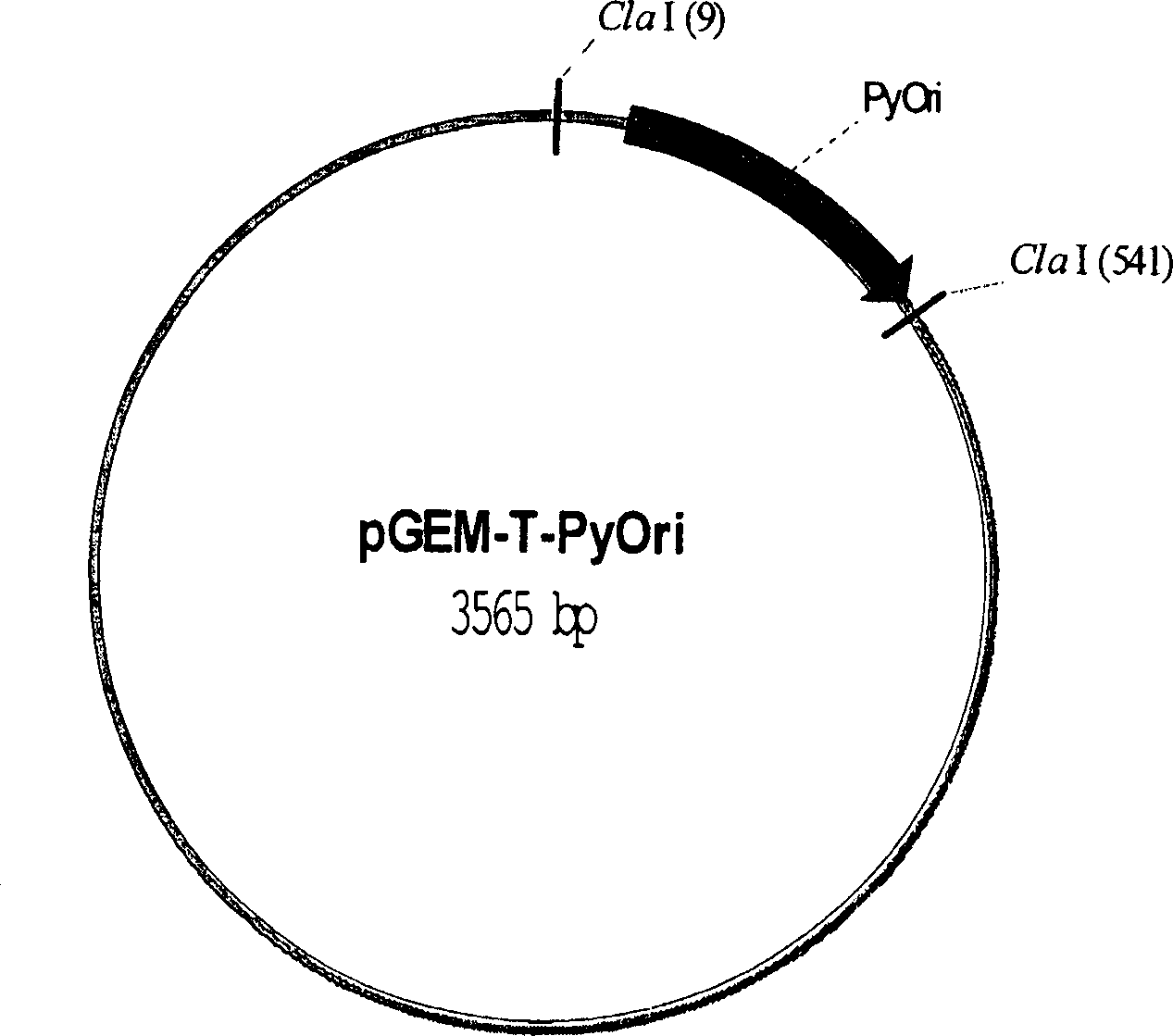
[0125] The expression vector pPyOriLT ( Figure 1G ) into CHO cells to demonstrate episomal replication. Thus, pPyOriLT contains PyOri and encodes PyLT, two essential viral elements, and is sufficient to initiate plasmid DNA replication in the presence of permissive CHO cytokines. Monitor plasmid DNA replication for 3 days. Purify low-molecular-weight DNA from transfected cells according to the Hirt extraction technique described above ( Image 6 (1)). Plasmid replication is detected by resistance to DpnI, which cleaves only when its recognition site is methylated. DNA purified from E. coli dam+ strains (lanes 1 to 4) is a substrate for DpnI (lane 5), whereas plasmid DNA undergoing one or more rounds of DNA replication in mammalian cells is resistant to DpnI cleavage (lanes 11-20) . as in Image 6 As shown in (2), the total amount of non-replicating DNA (Dpnl sensitive) dropped rapidly from 7000 copies per cell to less than 100...
Embodiment 3
[0127] (a) Generation of CHO-T cell lines
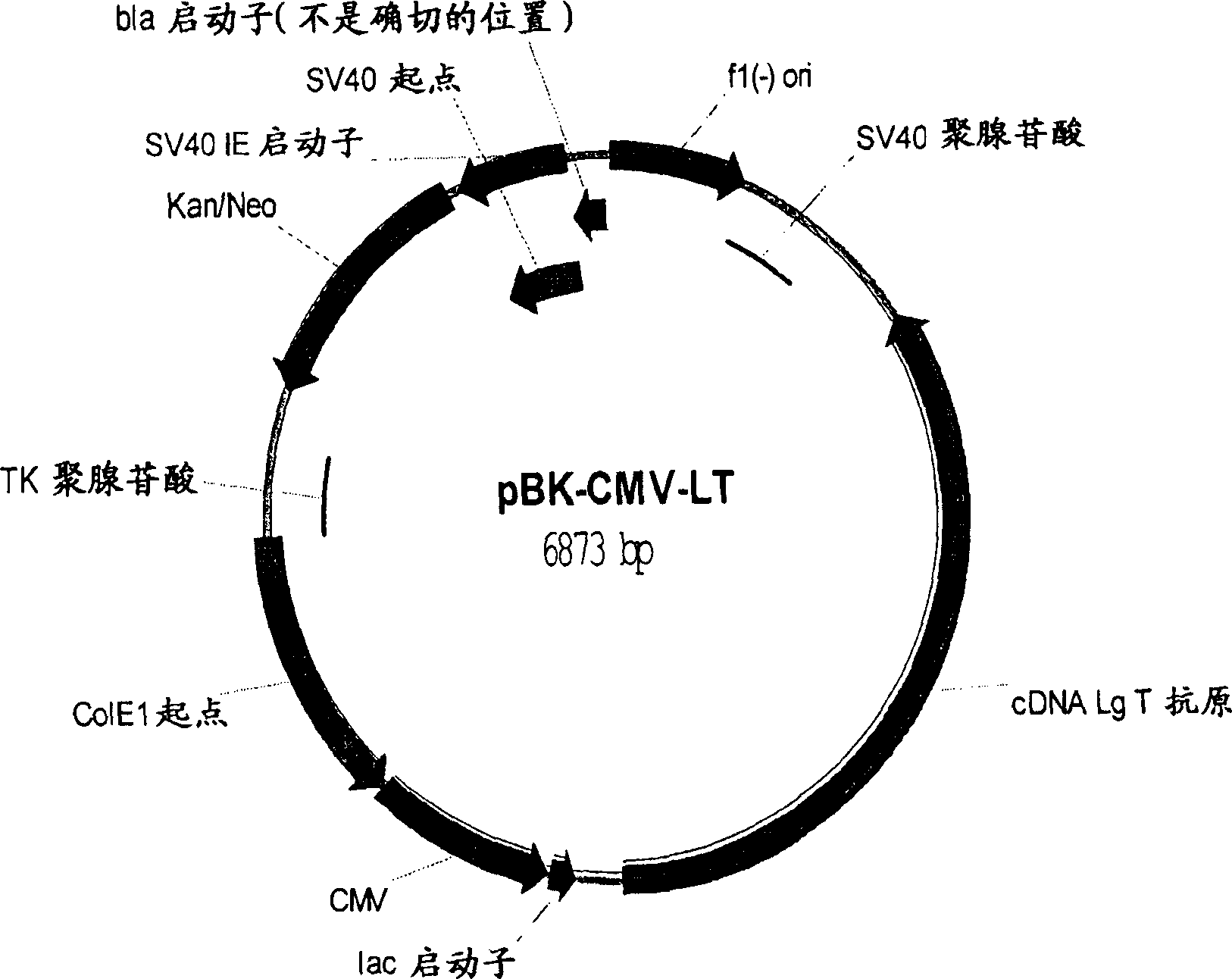
[0128]The cDNA for the Py large T antigen was obtained from ATCC and cloned into the expression vector pBK-CMV (Clonetech). CHO-K1 (ATCC, CCL61) was transfected with the expression plasmid pBK-CMV-LT (Figure 1). Cells were selected in G418 (400 μg / ml) for two weeks. by as in figure 2 The immunofluorescence staining shown in further confirmed the expression of Py large T antigen. Single cells were isolated by fluorescence-activated cell sorting using a MoFlo cytometer (Dako-Cytomation, Fort Collins, Colorado, USA). separated as in image 3 Several clones with varying levels of large T antigens are shown. 21 clones with different large T antigen expression levels were expanded and Figure 4 Their ability to support DNA replication of a Py ori-containing plasmid (pNK-Ori-EGFP) was tested as shown in the Southern blot. pNK-Ori-EGFP was transfected into CHO-T clones. Low molecular weight DNA was extracted by the Hirt extraction me...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com