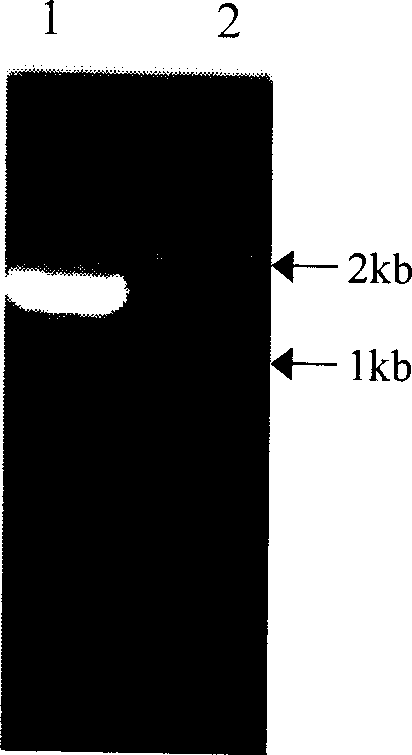
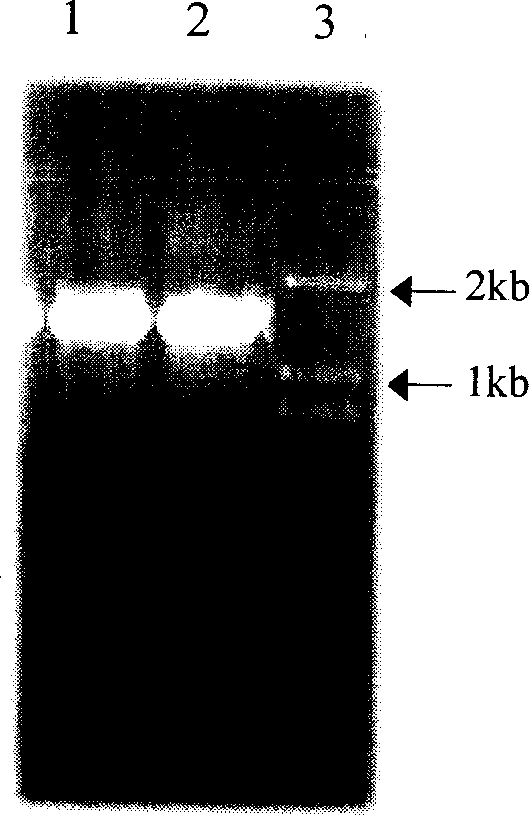
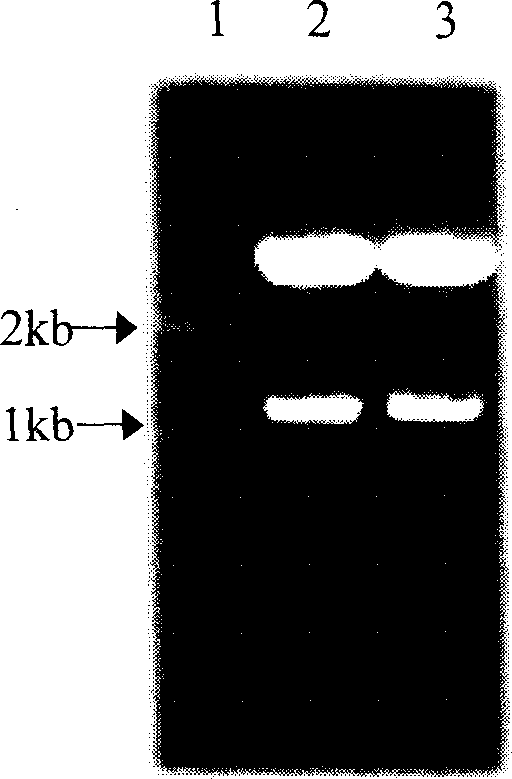
Medlar carotenoid synthase gene lycB and plasmid comprising the gene
A technology of carotene and synthesizing enzymes, applied in the field of genetic engineering, to achieve the effect of improving nutritional quality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Preparation of PCR template
[0019] Lycium barbarum cDNA library was provided by our laboratory, and λ phage DNA was extracted by liquid culture method.
[0020] Those skilled in the art can construct a wolfberry cDNA library in the laboratory. The construction steps of the library: (1) extraction of total RNA and purification of mRNA; (2) synthesis of cDNA double strands; (3) connection of cDNA and adapter with ligase;
[0021] (4) Removal of small fragments and redundant adapters; (5) Cloning of cDNA into vectors; (6) Packaging of phages; (7) Quality inspection of cDNA libraries.
[0022] In the process of cDNA library construction, mRNA isolation kits, cDNA synthesis kits, LambdaDNA packaging kits, vector λExCell, etc. were used, all of which have been commercialized.
Embodiment 2
[0024] PCR amplification in vitro
[0025] A pair of primers were designed and synthesized according to the conserved sequences of LycB genes in citrus, Arabidopsis, pepper, tomato, tobacco, daffodil, etc.: Primer 1 (5′ end primer SEQ ID No.3) 5′-GGA TCC ATG GAT ACT TTA GTG AAA ACTCCA-3'; Primer 2 (3' end primer SEQ ID No. 4) 5'-GC GGC CGC TTA TTC TTT GTC CCG CAATAA GTT-3'. In the 25μl reaction system, add 10×PCR Buffer (Mg 2+ 1SmM) 2.5μl, dNTP (each 2.5mmol / L) 2μl, primer 1 (20μM) and primer 2 (20μM) each 1μl, Ex Taq DNA polymerase (5U / μl) 0.5μl, template DNA 1.5ng. Reaction conditions: 94°C 3min→(94°C 1min→59°C 1min→72°C 2min) 35 →72°C for 10 minutes. The amplification reaction was carried out on a German UNOII Biometra DNA expander. Electrophoresis on 1% agarose gel was performed after the reaction.
Embodiment 3
[0027] Cloning and DNA sequence determination of amplified products
[0028] The PCR amplified products were recovered using Baobiology DNA Fragment Recovery Kit. The recovered product was ligated with the pGEM-T vector under the action of T 4 DNA ligase, and the ligated product was transformed into Escherichia coli JM109. Blue-white selection was performed on ampicillin-containing media. White colonies were randomly picked, and a small amount of plasmid DNA was extracted by alkaline lysis, and identified by PCR and double enzyme digestion (SacI, NcoI). Sequence determination was performed by Huada Gene Shanghai Dingan Biotechnology Co., Ltd. using a DNA fluorescence automatic sequence analyzer.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com