Method of determining a chemotherapeutic regimen based on ERCCI expression
A gene expression and protocol technology, applied in the medical field, can solve problems such as failure to obtain
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 4
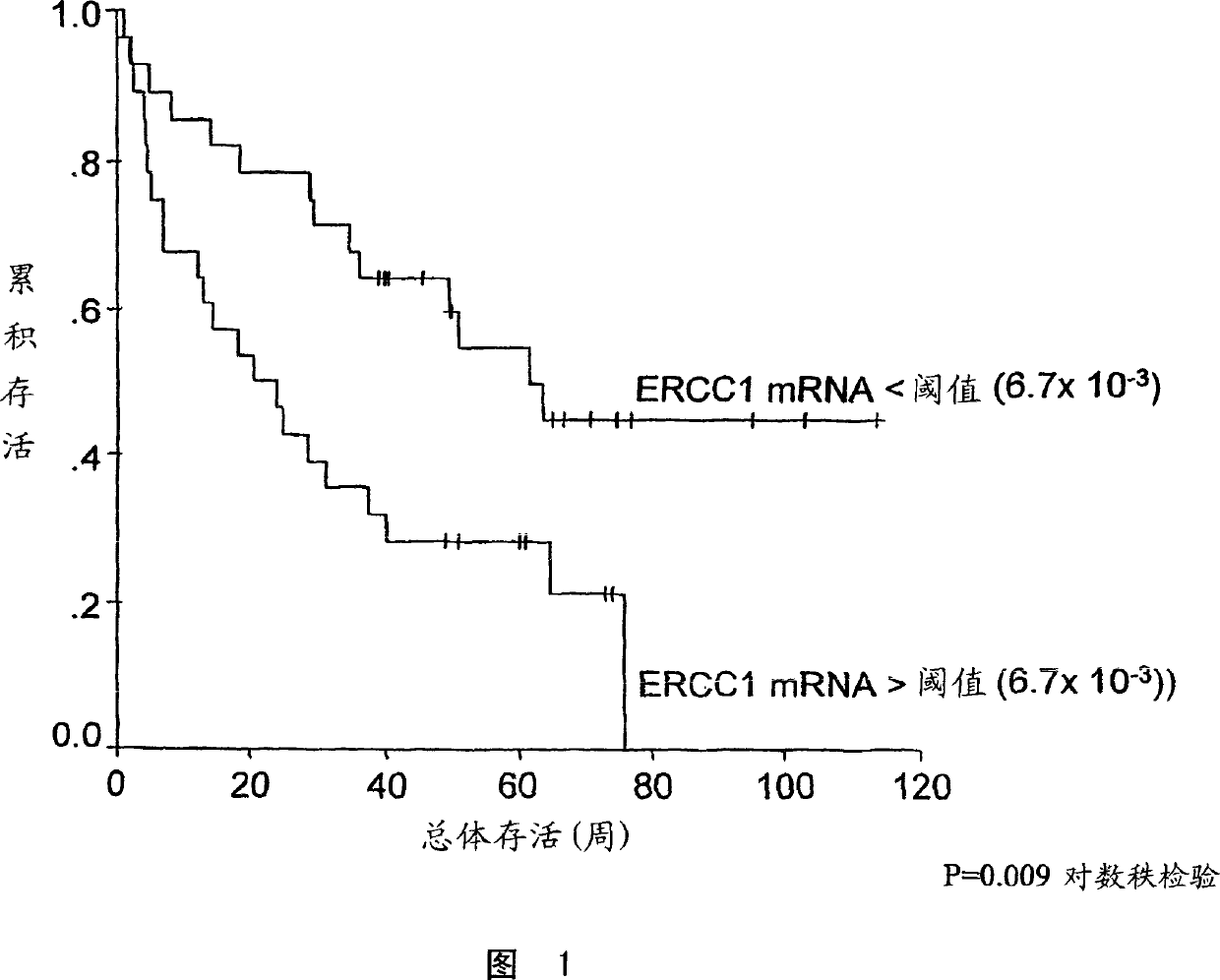
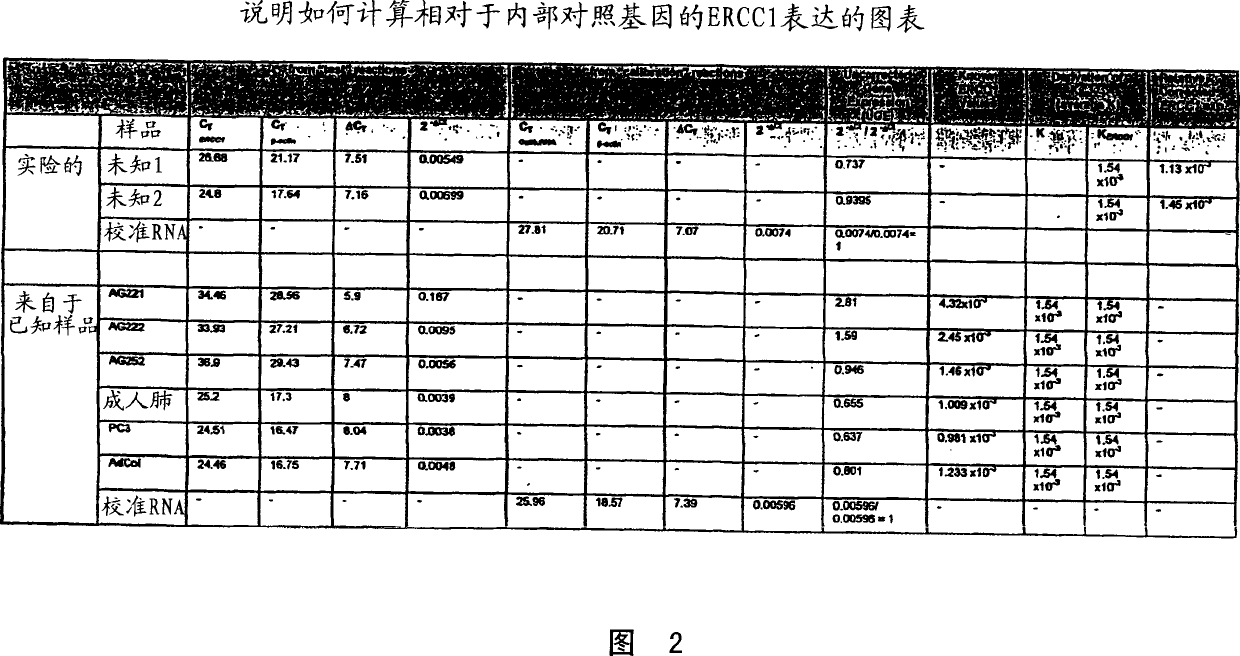
[0034] The "predetermined threshold level" is further defined as the corrected relative ERCC1 expression level of the tumor. Above this level, patients receiving platinum-based chemotherapy may have a low survival rate. When the corrected relative ERCC1 expression level of the tumor in patients receiving platinum-based chemotherapy regimens is lower than this domain level, it is associated with high patient survival rates. The domain correction expressed as the ratio of ERCC1:β-actin relative to ERCC1 expression is about 6.7×10 -3 . Figure 1, see Example 4. However, the present invention is not limited to the use of β-actin as an internal control.
[0035] When performing the method of this embodiment of the present invention, it is preferable to isolate the patient's tumor cells. Solid or lymphoma or part of it is removed from the patient by surgery or obtained by routine biopsy. RNA isolated from frozen or fresh tumor samples is extracted from cells by any typical method in the ...
Embodiment 1
[0076] Isolate RNA from FPE tissue
[0077] RNA is extracted from paraffin-embedded tissue through the following general process.
[0078] A. Deparaffinization and hydration of sections
[0079] (1) Put a part of the about 10 μM slice in a 1.5 mL plastic centrifuge tube.
[0080] (2) Add 600 μL of xylene, and shake the mixture vigorously at room temperature (about 20-25° C.) for about 10 minutes.
[0081] (3) At room temperature, centrifuge the sample for about 7 minutes at the maximum speed of the bench top centrifuge (about 10-20,000xg).
[0082] (4) Repeat steps 2 and 3 until most of the paraffin is dissolved. Depending on the amount of paraffin contained in the original sample part, it usually needs to be repeated 2 or more times.
[0083] (5) Use lower alcohol, preferably 100% ethanol (about 600 μL) to vigorously shake for about 3 minutes to remove the xylene solution.
[0084] (6) Centrifuge the test tube for about 7 minutes according to step (3). Pour out and discard the su...
Embodiment 2
[0097] mRNA reverse transcription and PCR
[0098]Reverse transcription: as exemplified in Example 1 and described in US Patent Application US09 / 469,338 filed on December 20, 1999 (incorporated here as a reference in its entirety), RNA is from microdissection or non-microdissection Isolated from formalin-fixed paraffin-embedded (FPE) tissue. After ethanol precipitation and centrifugation, the RNA pellets were dissolved in 50 μL of 5 mM Tris / Cl pH 8.0. M-MLV reverse transcriptase can extend an oligonucleotide primer that hybridizes to a single-stranded RNA or DNA template in the presence of deoxynucleotides to produce a complementary strand. The resulting RNA was reverse transcribed using M-MLV reverse transcriptase from Life Technologies and random hexamers. Reverse transcription is performed by mixing 25 μL of RNA solution with 25.5 μL of "Reverse Transcription Mix" (see below). Place the reactants in a thermal cycler, at 26°C for 8 minutes (for binding random hexamers to RNA), a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com