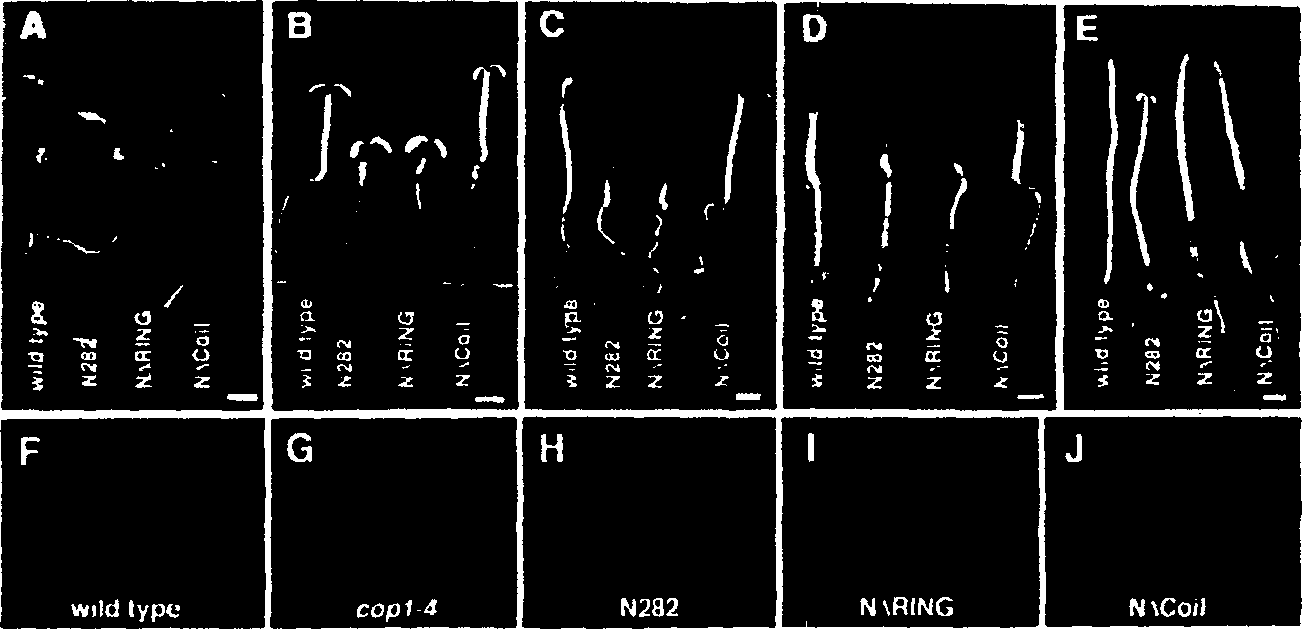
Plants and plants cells encoding wild type COP1 gene and coil domain thereof
A helical structure, plant cell technology, applied in angiosperms/flowering plants, botanical equipment and methods, biochemical equipment and methods, etc., can solve the impact, abnormal cotyledon growth and extension, seedling upward growth and unfavorable unearthed, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
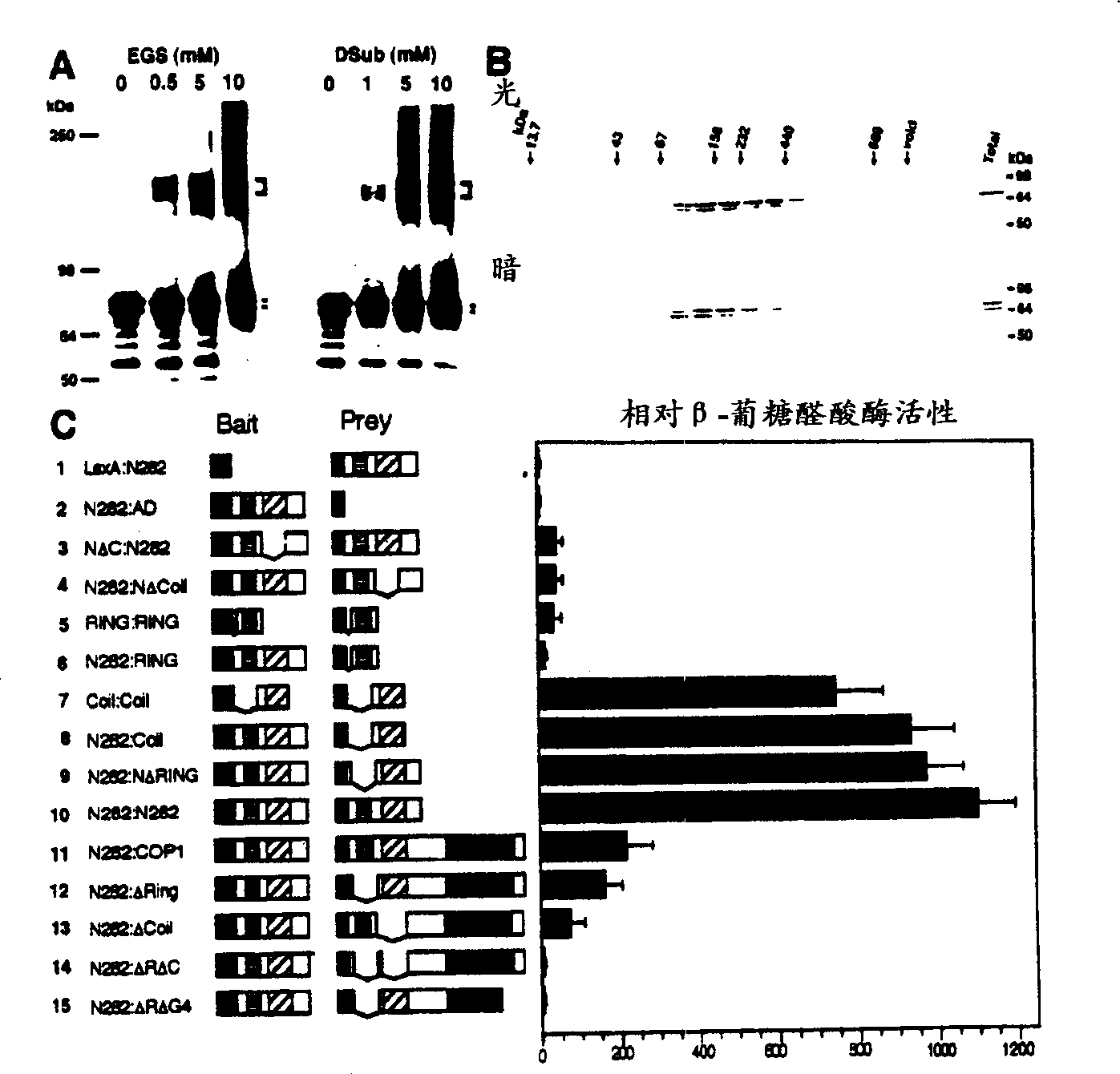
[0141] For gel filtration chromatography, 8 day old light or dark grown seedlings are homogenized with 2X gel filtration buffer, centrifuged at 4°C for 5 minutes, the buffer contains 50mM Tris-HCl (pH7.5), 440mM NaCl, 2mM MgCl 2 , 2μM ZnSO 4 0.5mM PMSF. A 25ml Superdex-200 FPLC column (Pharmacia) and 1X gel filtration buffer were used to separate the completely soluble protein at a rate of 0.5ml / min. All processes are carried out at 4°C, and for dark samples, it is also under green safety light. After the dead volume (7.5ml), each successive 0.5ml fraction was collected, concentrated and subjected to 10% SDS-PAGE, followed by protein immunohybridization analysis. The molecular weight standards used to estimate the size of natural COP1 protein are as follows: blue dextran (invalid); thyroglobulin (669kDa); ferritin (440kDa); catalase (232kDa); aldolase (158kDa); BSA (67kDa); Ovalbumin (43kDa) and Ribonuclease A (13.7kDa) (Sigma). Example 1. COP1 forms dimers in vivo and in vitro
...
example 2
[0144] In order to further define the COP1 dimerization domain inside the COP1 N282 fragment, a series of deletion mutants of N282 were constructed and analyzed using the yeast two-hybrid system ( figure 1 C). The results indicate that the helical domain of COP1 is necessary for self-connection. Is also sufficient ( figure 1 C3, 1C4, 1C7 to 12C10). However, the lack of the helical domain on the N282 fragment can still produce weak and reproducible interactions, which are slightly stronger than the negative control (see figure 1 C1 to 1C4), indicating that there is residual COP1 self-association in other regions of the N-terminus. It seems that the ring finger domain is responsible for this residual activity, because this domain exhibits a weak interaction with itself ( figure 1 C5). The following results make it obvious that the ring finger in COP1 supports intramolecular linkage. In the complete C-terminal construct ( figure 1 C11 to 1C14), the deletion of the helical domain...
example 3
[0144] In order to further define the COP1 dimerization domain inside the COP1 N282 fragment, a series of deletion mutants of N282 were constructed and analyzed using the yeast two-hybrid system ( figure 1 C). The results indicate that the helical domain of COP1 is necessary for self-connection. Is also sufficient ( figure 1 C3, 1C4, 1C7 to 12C10). However, the lack of the helical domain on the N282 fragment can still produce weak and reproducible interactions, which are slightly stronger than the negative control (see figure 1 C1 to 1C4), indicating that there is residual COP1 self-association in other regions of the N-terminus. It seems that the ring finger domain is responsible for this residual activity, because this domain exhibits a weak interaction with itself ( figure 1 C5). The following results make it obvious that the ring finger in COP1 supports intramolecular linkage. In the complete C-terminal construct ( figure 1 C11 to 1C14), the deletion of the helical domain...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com